Profiling of alternative polyadenylation and gene expression in PEDV-infected IPEC-J2 cells
- PMID: 33620696
- PMCID: PMC7900649
- DOI: 10.1007/s11262-020-01817-6
Profiling of alternative polyadenylation and gene expression in PEDV-infected IPEC-J2 cells
Abstract
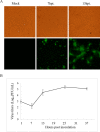
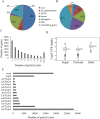
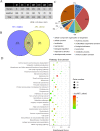
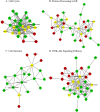
Since 2010, porcine epidemic diarrhea virus (PEDV) has received global attention with the emergence of variant strains characterized with high pathogenicity. The pathogen-host interaction after PEDV infection is still unclear. To investigate this issue, high-throughput-based sequencing technology is one of the optimal choices. In this study, we used in vitro transcription sequencing alternative polyadenylation sites (IVT-SAPAS) method, which allowed accurate profiling of gene expression and alternative polyadenylation (APA) sites to profile APA switching genes and differentially expressed genes (DEGs) in IPEC-J2 cells during PEDV variant strain infection. We found 804 APA switching genes, including switching in tandem 3' UTRs and switching between coding region and 3' UTR, and 1,677 DEGs in host after PEDV challenge. These genes participated in variety of biological processes such as cellular process, metabolism and immunity reactions. Moreover, 413 genes, most of which are the "focus" genes in interaction networks, were found to be involved in both APA switching genes and DEGs, suggesting these genes were synchronously regulated by different mechanisms. In summary, our results gave a relatively comprehensive insight into dynamic host-pathogen interactions in the regulation of host gene transcripts during PEDV infection.
Keywords: Alternative polyadenylation; Differential expression of host genes; Dynamic host–pathogen interactions; Porcine epidemic diarrhea virus.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

