Single-Cell RNA Sequencing Analysis of the Drosophila Larval Ventral Cord
- PMID: 33620770
- PMCID: PMC7942971
- DOI: 10.1002/cpz1.38
Single-Cell RNA Sequencing Analysis of the Drosophila Larval Ventral Cord
Erratum in
-
Group Correction Statement (Data Availability Statements).Curr Protoc. 2022 Aug;2(8):e552. doi: 10.1002/cpz1.552. Curr Protoc. 2022. PMID: 36005902 Free PMC article. No abstract available.
-
Group Correction Statement (Conflict of Interest Statements).Curr Protoc. 2022 Aug;2(8):e551. doi: 10.1002/cpz1.551. Curr Protoc. 2022. PMID: 36005903 Free PMC article. No abstract available.
Abstract
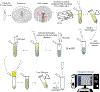
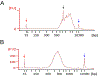
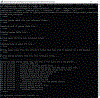
Drosophila provides a powerful genetic system and an excellent model to study the development and function of the nervous system. The fly's small brain and complex behavior has been instrumental in mapping neuronal circuits and elucidating the neural basis of behavior. The fast pace of fly development and the wealth of genetic tools has enabled systematic studies on cell differentiation and fate specification, and has uncovered strategies for axon guidance and targeting. The accessibility of neuronal structures and the ability to edit and manipulate gene expression in selective cells and/or synaptic compartments has revealed mechanisms for synapse assembly and neuronal connectivity. Recent advances in single-cell RNA sequencing (scRNA-seq) have further enhanced our appreciation and understanding of neuronal diversity in a fly brain. However, due to the small size of the fly brain and its constituent cells, scRNA-seq methodologies require a few adaptations. Here, we describe a set of protocols optimized for scRNA-seq analysis of the Drosophila larval ventral nerve cord, starting from tissue dissection and cell dissociation to cDNA library preparation, sequencing, and data analysis. We apply this workflow to three separate samples and detail the technical challenges associated with successful application of scRNA-seq to studies on neuronal diversity. An accompanying article (Vicidomini, Nguyen, Choudhury, Brody, & Serpe, 2021) presents a custom multistage analysis pipeline that integrates modules contained in different R packages to ensure high-flexibility, high-quality RNA-seq data analysis. These protocols are developed for Drosophila larval ventral nerve cord, but could easily be adapted to other tissues and model organisms. © 2021 U.S. Government. Basic Protocol 1: Dissection of larval ventral nerve cords and preparation of single-cell suspensions Basic Protocol 2: Preparation and sequencing of single-cell transcriptome libraries Basic Protocol 3: Alignment of raw sequencing data to indexed genome and generation of count matrices.
Keywords: 10× Genomics; Cell Ranger; Drosophila; cell dissociation; central nervous system; count matrix; demultiplexing; larval ventral nerve cord; reference genome; scRNA-seq.
Published 2021. This article is a U.S. Government work and is in the public domain in the USA.
Figures

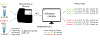


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

