Highly multiplexed 2-dimensional imaging mass cytometry analysis of HBV-infected liver
- PMID: 33621209
- PMCID: PMC8119221
- DOI: 10.1172/jci.insight.146883
Highly multiplexed 2-dimensional imaging mass cytometry analysis of HBV-infected liver
Abstract
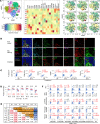
Studies of human hepatitis B virus (HBV) immune pathogenesis are hampered by limited access to liver tissues and technologies for detailed analyses. Here, utilizing imaging mass cytometry (IMC) to simultaneously detect 30 immune, viral, and structural markers in liver biopsies from patients with hepatitis B e antigen+ (HBeAg+) chronic hepatitis B, we provide potentially novel comprehensive visualization, quantitation, and phenotypic characterizations of hepatic adaptive and innate immune subsets that correlated with hepatocellular injury, histological fibrosis, and age. We further show marked correlations between adaptive and innate immune cell frequencies and phenotype, highlighting complex immune interactions within the hepatic microenvironment with relevance to HBV pathogenesis.
Keywords: Adaptive immunity; Fibrosis; Hepatology; Infectious disease; Innate immunity.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

