Transient non-specific DNA binding dominates the target search of bacterial DNA-binding proteins
- PMID: 33621478
- PMCID: PMC8022225
- DOI: 10.1016/j.molcel.2021.01.039
Transient non-specific DNA binding dominates the target search of bacterial DNA-binding proteins
Abstract
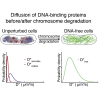
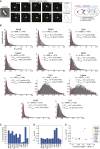
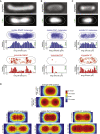
Despite their diverse biochemical characteristics and functions, all DNA-binding proteins share the ability to accurately locate their target sites among the vast excess of non-target DNA. Toward identifying universal mechanisms of the target search, we used single-molecule tracking of 11 diverse DNA-binding proteins in living Escherichia coli. The mobility of these proteins during the target search was dictated by DNA interactions rather than by their molecular weights. By generating cells devoid of all chromosomal DNA, we discovered that the nucleoid is not a physical barrier for protein diffusion but significantly slows the motion of DNA-binding proteins through frequent short-lived DNA interactions. The representative DNA-binding proteins (irrespective of their size, concentration, or function) spend the majority (58%-99%) of their search time bound to DNA and occupy as much as ∼30% of the chromosomal DNA at any time. Chromosome crowding likely has important implications for the function of all DNA-binding proteins.
Keywords: Chromosome-crowding; chromosome-free cells; single-molecule tracking; target search of bacterial DNA-binding proteins.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Bacia K., Kim S.A., Schwille P. Fluorescence cross-correlation spectroscopy in living cells. Nat. Methods. 2006;3:83–89. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

