CARMN Loss Regulates Smooth Muscle Cells and Accelerates Atherosclerosis in Mice
- PMID: 33622045
- PMCID: PMC7610708
- DOI: 10.1161/CIRCRESAHA.120.318688
CARMN Loss Regulates Smooth Muscle Cells and Accelerates Atherosclerosis in Mice
Abstract
[Figure: see text].
Keywords: atherosclerosis; cell proliferation; cholesterol; inflammation; phenotype.
Figures

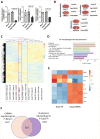


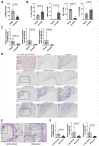
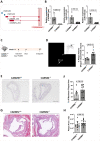
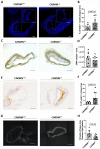

Comment in
-
Where in the (lncRNA) World Is CARMN?: Safeguarding Against Vascular Dysfunction.Circ Res. 2021 Apr 30;128(9):1276-1278. doi: 10.1161/CIRCRESAHA.121.319150. Epub 2021 Apr 29. Circ Res. 2021. PMID: 33914599 Free PMC article. No abstract available.
References
-
- Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

