Ectopic positioning of the cell division plane is associated with single amino acid substitutions in the FtsZ-recruiting SsgB in Streptomyces
- PMID: 33622102
- PMCID: PMC8061694
- DOI: 10.1098/rsob.200409
Ectopic positioning of the cell division plane is associated with single amino acid substitutions in the FtsZ-recruiting SsgB in Streptomyces
Abstract
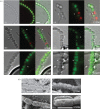
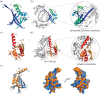
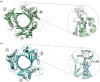
In most bacteria, cell division begins with the polymerization of the GTPase FtsZ at mid-cell, which recruits the division machinery to initiate cell constriction. In the filamentous bacterium Streptomyces, cell division is positively controlled by SsgB, which recruits FtsZ to the future septum sites and promotes Z-ring formation. Here, we show that various amino acid (aa) substitutions in the highly conserved SsgB protein result in ectopically placed septa that sever spores diagonally or along the long axis, perpendicular to the division plane. Fluorescence microscopy revealed that between 3.3% and 9.8% of the spores of strains expressing SsgB E120 variants were severed ectopically. Biochemical analysis of SsgB variant E120G revealed that its interaction with FtsZ had been maintained. The crystal structure of Streptomyces coelicolor SsgB was resolved and the key residues were mapped on the structure. Notably, residue substitutions (V115G, G118V, E120G) that are associated with septum misplacement localize in the α2-α3 loop region that links the final helix and the rest of the protein. Structural analyses and molecular simulation revealed that these residues are essential for maintaining the proper angle of helix α3. Our data suggest that besides altering FtsZ, aa substitutions in the FtsZ-recruiting protein SsgB also lead to diagonally or longitudinally divided cells in Streptomyces.
Keywords: SsgB; cell division; crystal structure; ectopic septa; mutagenesis.
Figures




Similar articles
-
SepG coordinates sporulation-specific cell division and nucleoid organization in Streptomyces coelicolor.Open Biol. 2016 Apr;6(4):150164. doi: 10.1098/rsob.150164. Open Biol. 2016. PMID: 27053678 Free PMC article.
-
Specific amino acid substitutions in β strand S2 of FtsZ cause spiraling septation and impair assembly cooperativity in Streptomyces spp.Mol Microbiol. 2019 Jul;112(1):184-198. doi: 10.1111/mmi.14262. Epub 2019 May 17. Mol Microbiol. 2019. PMID: 31002418
-
Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces.Genes Dev. 2011 Jan 1;25(1):89-99. doi: 10.1101/gad.600211. Genes Dev. 2011. PMID: 21205868 Free PMC article.
-
FtsZ-ring Architecture and Its Control by MinCD.Subcell Biochem. 2017;84:213-244. doi: 10.1007/978-3-319-53047-5_7. Subcell Biochem. 2017. PMID: 28500527 Review.
-
An intrinsically disordered linker plays a critical role in bacterial cell division.Semin Cell Dev Biol. 2015 Jan;37:3-10. doi: 10.1016/j.semcdb.2014.09.017. Epub 2014 Oct 13. Semin Cell Dev Biol. 2015. PMID: 25305578 Free PMC article. Review.
Cited by
-
Global Regulator AdpA_1075 Regulates Morphological Differentiation and Ansamitocin Production in Actinosynnema pretiosum subsp. auranticum.Bioengineering (Basel). 2022 Nov 21;9(11):719. doi: 10.3390/bioengineering9110719. Bioengineering (Basel). 2022. PMID: 36421120 Free PMC article.
-
Hyphal compartmentalization and sporulation in Streptomyces require the conserved cell division protein SepX.Nat Commun. 2022 Jan 10;13(1):71. doi: 10.1038/s41467-021-27638-1. Nat Commun. 2022. PMID: 35013186 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

