Identification of key pathways and genes in polycystic ovary syndrome via integrated bioinformatics analysis and prediction of small therapeutic molecules
- PMID: 33622336
- PMCID: PMC7901211
- DOI: 10.1186/s12958-021-00706-3
Identification of key pathways and genes in polycystic ovary syndrome via integrated bioinformatics analysis and prediction of small therapeutic molecules
Abstract
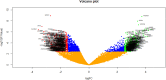
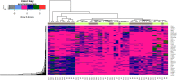
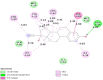
To enhance understanding of polycystic ovary syndrome (PCOS) at the molecular level; this investigation intends to examine the genes and pathways associated with PCOS by using an integrated bioinformatics analysis. Based on the expression profiling by high throughput sequencing data GSE84958 derived from the Gene Expression Omnibus (GEO) database, the differentially expressed genes (DEGs) between PCOS samples and normal controls were identified. We performed a functional enrichment analysis. A protein-protein interaction (PPI) network, miRNA- target genes and TF - target gene networks, were constructed and visualized, with which the hub gene nodes were identified. Validation of hub genes was performed by using receiver operating characteristic (ROC) and RT-PCR. Small drug molecules were predicted by using molecular docking. A total of 739 DEGs were identified, of which 360 genes were up regulated and 379 genes were down regulated. GO enrichment analysis revealed that up regulated genes were mainly involved in peptide metabolic process, organelle envelope and RNA binding and the down regulated genes were significantly enriched in plasma membrane bounded cell projection organization, neuron projection and DNA-binding transcription factor activity, RNA polymerase II-specific. REACTOME pathway enrichment analysis revealed that the up regulated genes were mainly enriched in translation and respiratory electron transport and the down regulated genes were mainly enriched in generic transcription pathway and transmembrane transport of small molecules. The top 10 hub genes (SAA1, ADCY6, POLR2K, RPS15, RPS15A, CTNND1, ESR1, NEDD4L, KNTC1 and NGFR) were identified from PPI network, miRNA - target gene network and TF - target gene network. The modules analysis showed that genes in modules were mainly associated with the transport of respiratory electrons and signaling NGF, respectively. We find a series of crucial genes along with the pathways that were most closely related with PCOS initiation and advancement. Our investigations provide a more detailed molecular mechanism for the progression of PCOS, detail information on the potential biomarkers and therapeutic targets.
Keywords: biomarkers; differentially expressed gene; expression profiling by high throughput sequencing; pathway enrichment analysis; polycystic ovary syndrome.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
Identification of candidate biomarkers and therapeutic agents for heart failure by bioinformatics analysis.BMC Cardiovasc Disord. 2021 Jul 4;21(1):329. doi: 10.1186/s12872-021-02146-8. BMC Cardiovasc Disord. 2021. PMID: 34218797 Free PMC article.
-
Investigation of candidate genes and mechanisms underlying obesity associated type 2 diabetes mellitus using bioinformatics analysis and screening of small drug molecules.BMC Endocr Disord. 2021 Apr 26;21(1):80. doi: 10.1186/s12902-021-00718-5. BMC Endocr Disord. 2021. PMID: 33902539 Free PMC article.
-
Pathway and network-based analysis of genome-wide association studies and RT-PCR validation in polycystic ovary syndrome.Int J Mol Med. 2017 Nov;40(5):1385-1396. doi: 10.3892/ijmm.2017.3146. Epub 2017 Sep 20. Int J Mol Med. 2017. PMID: 28949383 Free PMC article.
-
Explore the potential molecular mechanism of polycystic ovarian syndrome by protein-protein interaction network analysis.Taiwan J Obstet Gynecol. 2021 Sep;60(5):807-815. doi: 10.1016/j.tjog.2021.07.005. Taiwan J Obstet Gynecol. 2021. PMID: 34507653 Review.
-
A Systematic Review and Integrated Bioinformatic Analysis of Candidate Genes and Pathways in the Endometrium of Patients With Polycystic Ovary Syndrome During the Implantation Window.Front Endocrinol (Lausanne). 2022 Jul 1;13:900767. doi: 10.3389/fendo.2022.900767. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35860699 Free PMC article.
Cited by
-
Systemic changes induced by autologous stem cell ovarian transplant in plasma proteome of women with impaired ovarian reserves.Aging (Albany NY). 2023 Dec 26;15(24):14553-14573. doi: 10.18632/aging.205400. Epub 2023 Dec 26. Aging (Albany NY). 2023. PMID: 38149997 Free PMC article.
-
Contribution of labor related gene subtype classification on heterogeneity of polycystic ovary syndrome.PLoS One. 2023 Mar 1;18(3):e0282292. doi: 10.1371/journal.pone.0282292. eCollection 2023. PLoS One. 2023. PMID: 36857354 Free PMC article.
-
Identification of mitophagy-related biomarkers and immune infiltration in polycystic ovarian syndrome by bioinformatic study.Heliyon. 2025 Jan 23;11(3):e42202. doi: 10.1016/j.heliyon.2025.e42202. eCollection 2025 Feb 15. Heliyon. 2025. PMID: 39916834 Free PMC article.
-
The impact of GABA and GABAergic pathway in polycystic ovary syndrome: a systematic review.Obstet Gynecol Sci. 2025 Mar;68(2):93-108. doi: 10.5468/ogs.24255. Epub 2025 Feb 11. Obstet Gynecol Sci. 2025. PMID: 39935052 Free PMC article.
-
Never a dull enzyme, RNA polymerase II.Transcription. 2023 Nov;14(1-2):49-67. doi: 10.1080/21541264.2023.2208023. Epub 2023 May 2. Transcription. 2023. PMID: 37132022 Free PMC article. Review.
References
-
- Belenkaia LV, Lazareva LM, Walker W, Lizneva DV, Suturina LV. Criteria, phenotypes and prevalence of polycystic ovary syndrome. Minerva Ginecol 2019;71(3):211-223. doi:10.23736/S0026-4784.19.04404-6 - PubMed
-
- Oliver-Williams C, Vassard D, Pinborg A, Schmidt L. Risk of cardiovascular disease for women with polycystic ovary syndrome: results from a national Danish registry cohort study. Eur J Prev Cardiol. 2020:2047487320939674. 10.1177/2047487320939674. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

