Molecular Features of Cancer-associated Fibroblast Subtypes and their Implication on Cancer Pathogenesis, Prognosis, and Immunotherapy Resistance
- PMID: 33622705
- PMCID: PMC8102353
- DOI: 10.1158/1078-0432.CCR-20-4226
Molecular Features of Cancer-associated Fibroblast Subtypes and their Implication on Cancer Pathogenesis, Prognosis, and Immunotherapy Resistance
Abstract
Purpose: Cancer-associated fibroblasts (CAFs) are an important component of the tumor microenvironment, but a systematic investigation of their molecular characteristics and clinical relevance are lacking. Here, we sought to compare CAFs across multiple cancer types to identify critical molecular pathways activated in CAF subtypes, which may contribute to clinical outcome, disease progression, and immunotherapy resistance.
Experimental design: We performed integrated analysis of CAFs from melanoma, head and neck squamous cell carcinoma, and lung cancer, and identified the molecular characteristics that are distinctly active in each CAF subtype. Gene signatures for individual CAF subtypes were identified and used to study the association of subtype abundance with clinical outcome and immunotherapy resistance.
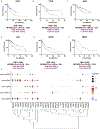
Results: We identified six CAF subtypes (pan-CAF) shared across cancer types and uncovered the molecular characteristics and genetic pathways distinguishing them. Interestingly, these CAF subtypes express distinct immunosuppressive factors, such as CXCL12 and CXLC14, and stem cell-promoting factor IL6. In addition, we identified novel transcriptional drivers (MEF2C, TWIST1, NR1H3, RELB, and FOXM1) key to CAF heterogeneity. Furthermore, we showed that CAF subtypes were associated with different clinical outcomes and uncovered key molecular pathways that could activate or suppress cancer progression or were involved in resistance to anti-PD1 or anti-PD-L1 immunotherapy.
Conclusions: Our study identifies the molecular characteristics of CAF subtypes shared across several cancer types, implicates cancer types that may benefit from CAF subtype targeted therapies, and identifies specific CAF subtypes associated with immunotherapy resistance.
©2021 American Association for Cancer Research.
Conflict of interest statement
Figures
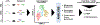




References
-
- Chen X and Song E. Turning foes to friends: targeting cancer-associated fibroblasts. Nat Rev Drug Discov 2019;18:99–115. - PubMed
-
- Su S, Chen J, Yao H, Liu J, Yu S, Lao L, et al. CD10+ GPR77+ cancer-associated fibroblasts promote cancer formation and chemoresistance by sustaining cancer stemness. Cell 2018;172:841–856. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

