Local computational methods to improve the interpretability and analysis of cryo-EM maps
- PMID: 33623015
- PMCID: PMC7902670
- DOI: 10.1038/s41467-021-21509-5
Local computational methods to improve the interpretability and analysis of cryo-EM maps
Abstract
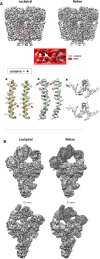
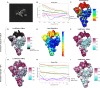
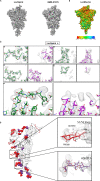
Cryo-electron microscopy (cryo-EM) maps usually show heterogeneous distributions of B-factors and electron density occupancies and are typically B-factor sharpened to improve their contrast and interpretability at high-resolutions. However, 'over-sharpening' due to the application of a single global B-factor can distort processed maps causing connected densities to appear broken and disconnected. This issue limits the interpretability of cryo-EM maps, i.e. ab initio modelling. In this work, we propose 1) approaches to enhance high-resolution features of cryo-EM maps, while preventing map distortions and 2) methods to obtain local B-factors and electron density occupancy maps. These algorithms have as common link the use of the spiral phase transformation and are called LocSpiral, LocBSharpen, LocBFactor and LocOccupancy. Our results, which include improved maps of recent SARS-CoV-2 structures, show that our methods can improve the interpretability and analysis of obtained reconstructions.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Wandzik, J. M. et al. A structure-based model for the complete transcription cycle of influenza polymerase. Cell10.1016/j.cell.2020.03.061 (2020). - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

