Prioritizing variants of uncertain significance for reclassification using a rule-based algorithm in inherited retinal dystrophies
- PMID: 33623043
- PMCID: PMC7902814
- DOI: 10.1038/s41525-021-00182-z
Prioritizing variants of uncertain significance for reclassification using a rule-based algorithm in inherited retinal dystrophies
Abstract
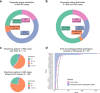
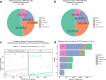
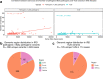
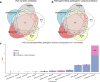
Inherited retinal dystrophies (IRD) are a highly heterogeneous group of rare diseases with a molecular diagnostic rate of >50%. Reclassification of variants of uncertain significance (VUS) poses a challenge for IRD diagnosis. We collected 668 IRD cases analyzed by our geneticists using two different clinical exome-sequencing tests. We identified 114 unsolved cases pending reclassification of 125 VUS and studied their genomic, functional, and laboratory-specific features, comparing them to pathogenic and likely pathogenic variants from the same cohort (N = 390). While the clinical exome used did not show differences in diagnostic rate, the more IRD-experienced geneticist reported more VUS (p = 4.07e-04). Significantly fewer VUS were reported in recessive cases (p = 2.14e-04) compared to other inheritance patterns, and of all the genes analyzed, ABCA4 and IMPG2 had the lowest and highest VUS frequencies, respectively (p = 3.89e-04, p = 6.93e-03). Moreover, few frameshift and stop-gain variants were found to be informed VUS (p = 6.73e-08 and p = 2.93e-06). Last, we applied five pathogenicity predictors and found there is a significant proof of deleteriousness when all score for pathogenicity in missense variants. Altogether, these results provided input for a set of rules that correctly reclassified ~70% of VUS as pathogenic in validation datasets. Disease- and setting-specific features influence VUS reporting. Comparison with pathogenic and likely pathogenic variants can prioritize VUS more likely to be reclassified as causal.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Martin-Merida I, et al. Toward the mutational landscape of autosomal dominant retinitis pigmentosa: a comprehensive analysis of 258 Spanish families. Investig. Ophthalmol. Vis. Sci. 2018;59:2345–2354. - PubMed
-
- Martin-Merida, I. et al. Genomic landscape of sporadic retinitis pigmentosa. ophthalmology 10.1016/j.ophtha.2019.03.018. (2019). - PubMed
Grants and funding
- PEJ-2017-AI/BMD7256/Comunidad de Madrid
- B2017/BMD-3721/Comunidad de Madrid
- FI17/00192/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- CP12/03256/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- CP16/00116/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
LinkOut - more resources
Full Text Sources
Other Literature Sources

