Engineering organoids
- PMID: 33623712
- PMCID: PMC7893133
- DOI: 10.1038/s41578-021-00279-y
Engineering organoids
Abstract
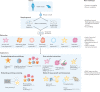
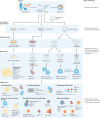
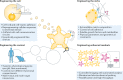
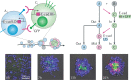
Organoids are in vitro miniaturized and simplified model systems of organs that have gained enormous interest for modelling tissue development and disease, and for personalized medicine, drug screening and cell therapy. Despite considerable success in culturing physiologically relevant organoids, challenges remain to achieve real-life applications. In particular, the high variability of self-organizing growth and restricted experimental and analytical access hamper the translatability of organoid systems. In this Review, we argue that many limitations of traditional organoid culture can be addressed by engineering approaches at all levels of organoid systems. We investigate cell surface and genetic engineering approaches, and discuss stem cell niche engineering based on the design of matrices that allow spatiotemporal control of organoid growth and shape-guided morphogenesis. We examine how microfluidic approaches and lessons learnt from organs-on-a-chip enable the integration of mechano-physiological parameters and increase accessibility of organoids to improve functional readouts. Applying engineering principles to organoids increases reproducibility and provides experimental control, which will, ultimately, be required to enable clinical translation.
Keywords: Morphogenesis; Organogenesis; Stem cells; Tissue engineering.
© Springer Nature Limited 2021.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
