Characterizing genomic variants and mutations in SARS-CoV-2 proteins from Indian isolates
- PMID: 33623833
- PMCID: PMC7893251
- DOI: 10.1016/j.genrep.2021.101044
Characterizing genomic variants and mutations in SARS-CoV-2 proteins from Indian isolates
Abstract
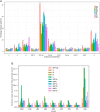
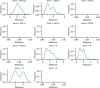
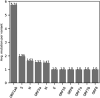
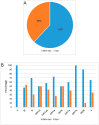
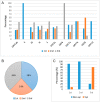
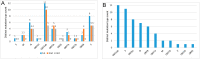
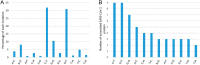
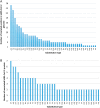
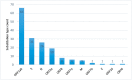
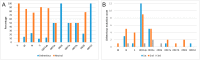
SARS-CoV-2 is mutating and creating divergent variants by altering the composition of essential constituent proteins. Pharmacologically, it is crucial to understand the diverse mechanism of mutations for stable vaccine or anti-viral drug design. Our current study concentrates on all the constituent proteins of 469 SARS-CoV-2 genome samples, derived from Indian patients. However, the study may easily be extended to the samples across the globe. We perform clustering analysis towards identifying unique variants in each of the SARS-CoV-2 proteins. A total of 536 mutated positions within the coding regions of SARS-CoV-2 proteins are detected among the identified variants from Indian isolates. We quantify mutations by focusing on the unique variants of each SARS-CoV-2 protein. We report the average number of mutation per variant, percentage of mutated positions, synonymous and non-synonymous mutations, mutations occurring in three codon positions and so on. Our study reveals the most susceptible six (06) proteins, which are ORF1ab, Spike (S), Nucleocapsid (N), ORF3a, ORF7a, and ORF8. Several non-synonymous substitutions are observed to be unique in different SARS-CoV-2 proteins. A total of 57 possible deleterious amino acid substitutions are predicted, which may impact on the protein functions. Several mutations show a large decrease in protein stability and are observed in putative functional domains of the proteins that might have some role in disease pathogenesis. We observe a good number of physicochemical property change during above deleterious substitutions.
Keywords: AA, amino acid; COVID-19; CP, codon position; CTD, C-terminal domain; CoV, coronaviruses; Codon position; Deleterious substitutions; Functional domain; HR, heptapeptide repeat; Mut, mutation; NS, non-synonymous; NTD, N-terminal domain; Non-synonymous mutations; Protein stability; SARS, severe acute respiratory syndrome; Syn, synonymous; TM, transmembrane domain.
© 2021 Published by Elsevier Inc.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Genomic Surveillance and Mutation Analysis of SARS-CoV-2 Variants among Patients in Saudi Arabia.Microorganisms. 2024 Feb 26;12(3):467. doi: 10.3390/microorganisms12030467. Microorganisms. 2024. PMID: 38543518 Free PMC article.
-
A study on non-synonymous mutational patterns in structural proteins of SARS-CoV-2.Genome. 2021 Jul;64(7):665-678. doi: 10.1139/gen-2020-0157. Epub 2021 Mar 31. Genome. 2021. PMID: 33788636
-
The status and analysis of common mutations found in the SARS-CoV-2 whole genome sequences from Bangladesh.Gene Rep. 2022 Jun;27:101608. doi: 10.1016/j.genrep.2022.101608. Epub 2022 Apr 4. Gene Rep. 2022. PMID: 35399222 Free PMC article.
-
Effect of Genomic and Amino Acid Sequence Mutation on Virulence and Therapeutic Target of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS COV-2).Infect Drug Resist. 2021 Jun 14;14:2187-2192. doi: 10.2147/IDR.S307374. eCollection 2021. Infect Drug Resist. 2021. PMID: 34163183 Free PMC article. Review.
-
Characterization of SARS-CoV-2 different variants and related morbidity and mortality: a systematic review.Eur J Med Res. 2021 Jun 8;26(1):51. doi: 10.1186/s40001-021-00524-8. Eur J Med Res. 2021. PMID: 34103090 Free PMC article.
Cited by
-
Wastewater sequencing from a rural community enables identification of widespread adaptive mutations in a SARS-CoV-2 alpha variant.Sci Rep. 2025 May 28;15(1):18657. doi: 10.1038/s41598-025-03771-5. Sci Rep. 2025. PMID: 40437225 Free PMC article.
-
The Algerian Chapter of SARS-CoV-2 Pandemic: An Evolutionary, Genetic, and Epidemiological Prospect.Viruses. 2021 Aug 2;13(8):1525. doi: 10.3390/v13081525. Viruses. 2021. PMID: 34452390 Free PMC article.
-
Genomic Surveillance and Mutation Analysis of SARS-CoV-2 Variants among Patients in Saudi Arabia.Microorganisms. 2024 Feb 26;12(3):467. doi: 10.3390/microorganisms12030467. Microorganisms. 2024. PMID: 38543518 Free PMC article.
-
Comparative analysis of human coronaviruses focusing on nucleotide variability and synonymous codon usage patterns.Genomics. 2021 Jul;113(4):2177-2188. doi: 10.1016/j.ygeno.2021.05.008. Epub 2021 May 19. Genomics. 2021. PMID: 34019999 Free PMC article.
-
Intestinal viral infections of nSARS-CoV2 in the Indian community: Risk of virus spread in India.J Med Virol. 2022 Apr;94(4):1315-1329. doi: 10.1002/jmv.27480. Epub 2021 Dec 3. J Med Virol. 2022. PMID: 34825708 Free PMC article. Review.
References
-
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3clpro) structure: basis for design of anti-sars drugs. Science. 2003;300:1763–1767. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
