The NAC transcription factor FaRIF controls fruit ripening in strawberry
- PMID: 33624824
- PMCID: PMC8254488
- DOI: 10.1093/plcell/koab070
The NAC transcription factor FaRIF controls fruit ripening in strawberry
Abstract
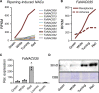
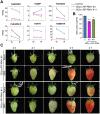
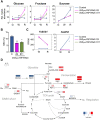
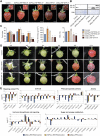
In contrast to climacteric fruits such as tomato, the knowledge on key regulatory genes controlling the ripening of strawberry, a nonclimacteric fruit, is still limited. NAC transcription factors (TFs) mediate different developmental processes in plants. Here, we identified and characterized Ripening Inducing Factor (FaRIF), a NAC TF that is highly expressed and induced in strawberry receptacles during ripening. Functional analyses based on stable transgenic lines aimed at silencing FaRIF by RNA interference, either from a constitutive promoter or the ripe receptacle-specific EXP2 promoter, as well as overexpression lines showed that FaRIF controls critical ripening-related processes such as fruit softening and pigment and sugar accumulation. Physiological, metabolome, and transcriptome analyses of receptacles of FaRIF-silenced and overexpression lines point to FaRIF as a key regulator of strawberry fruit ripening from early developmental stages, controlling abscisic acid biosynthesis and signaling, cell-wall degradation, and modification, the phenylpropanoid pathway, volatiles production, and the balance of the aerobic/anaerobic metabolism. FaRIF is therefore a target to be modified/edited to control the quality of strawberry fruits.
© The Author(s) 2021. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Figures



Similar articles
-
MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria x ananassa fruits.J Exp Bot. 2014 Feb;65(2):401-17. doi: 10.1093/jxb/ert377. Epub 2013 Nov 25. J Exp Bot. 2014. PMID: 24277278
-
A FvERF3-FvNAC073 module regulates strawberry fruit size and ripening.Plant J. 2025 Jun;122(5):e70262. doi: 10.1111/tpj.70262. Plant J. 2025. PMID: 40504991
-
Analysis of eight phytohormone concentrations, expression levels of ABA biosynthesis genes, and ripening-related transcription factors during fruit development in strawberry.J Plant Physiol. 2019 Aug;239:52-60. doi: 10.1016/j.jplph.2019.05.013. Epub 2019 Jun 3. J Plant Physiol. 2019. PMID: 31185317
-
Abscisic acid perception and signaling transduction in strawberry: a model for non-climacteric fruit ripening.Plant Signal Behav. 2011 Dec;6(12):1950-3. doi: 10.4161/psb.6.12.18024. Plant Signal Behav. 2011. PMID: 22095148 Free PMC article. Review.
-
The interplay between ABA/ethylene and NAC TFs in tomato fruit ripening: a review.Plant Mol Biol. 2021 Jun;106(3):223-238. doi: 10.1007/s11103-021-01128-w. Epub 2021 Feb 25. Plant Mol Biol. 2021. PMID: 33634368 Review.
Cited by
-
FvWRKY48 binds to the pectate lyase FvPLA promoter to control fruit softening in Fragaria vesca.Plant Physiol. 2022 Jun 1;189(2):1037-1049. doi: 10.1093/plphys/kiac091. Plant Physiol. 2022. PMID: 35238391 Free PMC article.
-
Autophagy Is Required for Strawberry Fruit Ripening.Front Plant Sci. 2021 Aug 27;12:688481. doi: 10.3389/fpls.2021.688481. eCollection 2021. Front Plant Sci. 2021. PMID: 34512686 Free PMC article.
-
The synthesis of papaya fruit flavor-related linalool was regulated by CpTPS18 and CpNAC56.Plant Reprod. 2024 Sep;37(3):295-308. doi: 10.1007/s00497-023-00486-3. Epub 2023 Nov 15. Plant Reprod. 2024. PMID: 37966580
-
Comparative genomic analysis and phylogeny of NAC25 gene from cultivated and wild Coffea species.Front Plant Sci. 2022 Sep 16;13:1009733. doi: 10.3389/fpls.2022.1009733. eCollection 2022. Front Plant Sci. 2022. PMID: 36186041 Free PMC article.
-
Roles of abscisic acid in regulating ripening and quality of strawberry, a model non-climacteric fruit.Hortic Res. 2022 Apr 22;9:uhac089. doi: 10.1093/hr/uhac089. eCollection 2022. Hortic Res. 2022. PMID: 35795383 Free PMC article.
References
-
- Aharoni A, De Vos CH, Wein M, Sun Z, Greco R, Kroon A, Mol JN, O’Connell AP (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J 28: 319–332 - PubMed
-
- Barceló M, Mansouri ElI, Mercado J-Á, Quesada MA, Pliego-Alfaro F (1998) Regeneration and transformation via Agrobacterium tumefaciens of the strawberry cultivar Chandler. Plant Cell Tissue Organ Culture 54: 29–36
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

