Sharing ICU Patient Data Responsibly Under the Society of Critical Care Medicine/European Society of Intensive Care Medicine Joint Data Science Collaboration: The Amsterdam University Medical Centers Database (AmsterdamUMCdb) Example
- PMID: 33625129
- PMCID: PMC8132908
- DOI: 10.1097/CCM.0000000000004916
Sharing ICU Patient Data Responsibly Under the Society of Critical Care Medicine/European Society of Intensive Care Medicine Joint Data Science Collaboration: The Amsterdam University Medical Centers Database (AmsterdamUMCdb) Example
Abstract
Objectives: Critical care medicine is a natural environment for machine learning approaches to improve outcomes for critically ill patients as admissions to ICUs generate vast amounts of data. However, technical, legal, ethical, and privacy concerns have so far limited the critical care medicine community from making these data readily available. The Society of Critical Care Medicine and the European Society of Intensive Care Medicine have identified ICU patient data sharing as one of the priorities under their Joint Data Science Collaboration. To encourage ICUs worldwide to share their patient data responsibly, we now describe the development and release of Amsterdam University Medical Centers Database (AmsterdamUMCdb), the first freely available critical care database in full compliance with privacy laws from both the United States and Europe, as an example of the feasibility of sharing complex critical care data.
Setting: University hospital ICU.
Subjects: Data from ICU patients admitted between 2003 and 2016.
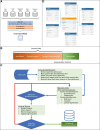
Interventions: We used a risk-based deidentification strategy to maintain data utility while preserving privacy. In addition, we implemented contractual and governance processes, and a communication strategy. Patient organizations, supporting hospitals, and experts on ethics and privacy audited these processes and the database.
Measurements and main results: AmsterdamUMCdb contains approximately 1 billion clinical data points from 23,106 admissions of 20,109 patients. The privacy audit concluded that reidentification is not reasonably likely, and AmsterdamUMCdb can therefore be considered as anonymous information, both in the context of the U.S. Health Insurance Portability and Accountability Act and the European General Data Protection Regulation. The ethics audit concluded that responsible data sharing imposes minimal burden, whereas the potential benefit is tremendous.
Conclusions: Technical, legal, ethical, and privacy challenges related to responsible data sharing can be addressed using a multidisciplinary approach. A risk-based deidentification strategy, that complies with both U.S. and European privacy regulations, should be the preferred approach to releasing ICU patient data. This supports the shared Society of Critical Care Medicine and European Society of Intensive Care Medicine vision to improve critical care outcomes through scientific inquiry of vast and combined ICU datasets.
Copyright © 2021 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the Society of Critical Care Medicine and Wolters Kluwer Health, Inc.
Conflict of interest statement
Dr. Sijbrands’ institution received funding from European Institute of Innovation and Technology (EIT) Health and Amgen. Drs. Kaplan and Bailey received funding from Society of Critical Care Medicine. Dr. Cecconi received funding from Directed Systems, Edwards Lifesciences, and Cheetah Medical. Dr. Churpek’s institution received funding from an EarlySense research grant; he is supported by National Institutes of Health (NIH) R01 (GM123193), and he has a patent pending for risk stratification algorithm for hospitalized patients (money from royalties from the University of Chicago). Dr. Clermont received funding from the NIH, Department of Defense, National Science Foundation, and NOMA AI. The remaining authors have disclosed that they do not have any potential conflicts of interest.
Figures


Comment in
-
Reidentification of Protected Health Information: Can the Risk Be Quantified?Crit Care Med. 2021 Jun 1;49(6):1003-1006. doi: 10.1097/CCM.0000000000004931. Crit Care Med. 2021. PMID: 34011836 No abstract available.
References
-
- Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med. 2019; 380:1347–1358 - PubMed
-
- Beam AL, Kohane IS. Big data and machine learning in health care. JAMA. 2018; 319:1317–1318 - PubMed
-
- Bailly S, Meyfroidt G, Timsit J-F. What’s new in ICU in 2050: Big data and machine learning. Intensive Care Med. 2017; 44:1524–1527 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

