The state of oligomerization of Rubisco controls the rate of synthesis of the Rubisco large subunit in Chlamydomonas reinhardtii
- PMID: 33625514
- PMCID: PMC8254502
- DOI: 10.1093/plcell/koab061
The state of oligomerization of Rubisco controls the rate of synthesis of the Rubisco large subunit in Chlamydomonas reinhardtii
Abstract
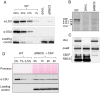
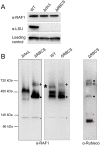
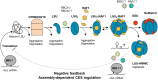
Ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) is present in all photosynthetic organisms and is a key enzyme for photosynthesis-driven life on Earth. Its most prominent form is a hetero-oligomer in which small subunits (SSU) stabilize the core of the enzyme built from large subunits (LSU), yielding, after a chaperone-assisted multistep assembly process, an LSU8SSU8 hexadecameric holoenzyme. Here we use Chlamydomonas reinhardtii and a combination of site-directed mutants to dissect the multistep biogenesis pathway of Rubisco in vivo. We identify assembly intermediates, in two of which LSU are associated with the RAF1 chaperone. Using genetic and biochemical approaches we further unravel a major regulation process during Rubisco biogenesis, in which LSU translation is controlled by its ability to assemble with the SSU, via the mechanism of control by epistasy of synthesis (CES). Altogether this leads us to propose a model whereby the last assembly intermediate, an LSU8-RAF1 complex, provides the platform for SSU binding to form the Rubisco enzyme, and when SSU is not available, converts to a key regulatory form that exerts negative feedback on the initiation of LSU translation.
© The Author(s) 2021. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Figures










Similar articles
-
Subunit interface dynamics in hexadecameric rubisco.J Mol Biol. 2011 Sep 2;411(5):1083-98. doi: 10.1016/j.jmb.2011.06.052. Epub 2011 Jul 6. J Mol Biol. 2011. PMID: 21745478
-
Degradation of Rubisco SSU during oxidative stress triggers aggregation of Rubisco particles in Chlamydomonas reinhardtii.Planta. 2005 Nov;222(5):787-93. doi: 10.1007/s00425-005-0023-0. Epub 2005 Jul 15. Planta. 2005. PMID: 16025343
-
RNA binding activity of the ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit from Chlamydomonas reinhardtii.J Biol Chem. 2004 Mar 12;279(11):10148-56. doi: 10.1074/jbc.M308602200. Epub 2003 Dec 16. J Biol Chem. 2004. PMID: 14679208
-
Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase.Arch Biochem Biophys. 2003 Jun 15;414(2):141-9. doi: 10.1016/s0003-9861(03)00171-1. Arch Biochem Biophys. 2003. PMID: 12781765 Review.
-
Assembly-controlled regulation of chloroplast gene translation.Biochem Soc Trans. 2001 Aug;29(Pt 4):421-6. doi: 10.1042/bst0290421. Biochem Soc Trans. 2001. PMID: 11498001 Review.
Cited by
-
The chloroplast RNA-binding protein CP29A supports rbcL expression during cold acclimation.Proc Natl Acad Sci U S A. 2025 Feb 4;122(5):e2403969122. doi: 10.1073/pnas.2403969122. Epub 2025 Jan 29. Proc Natl Acad Sci U S A. 2025. PMID: 39879235 Free PMC article.
-
Rubisco feedback loop: control by epistasy of synthesis governs large subunit biosynthesis.Plant Cell. 2021 Jul 2;33(5):1407-1408. doi: 10.1093/plcell/koab066. Plant Cell. 2021. PMID: 35234954 Free PMC article. No abstract available.
-
Rubisco small subunit (RbCS) is co-opted by potyvirids as the scaffold protein in assembling a complex for viral intercellular movement.PLoS Pathog. 2024 Mar 4;20(3):e1012064. doi: 10.1371/journal.ppat.1012064. eCollection 2024 Mar. PLoS Pathog. 2024. PMID: 38437247 Free PMC article.
-
Assembly-dependent translational feedback regulation of photosynthetic proteins in land plants.Nat Plants. 2025 Aug 18. doi: 10.1038/s41477-025-02074-x. Online ahead of print. Nat Plants. 2025. PMID: 40825854
-
Plant organellar RNA maturation.Plant Cell. 2023 May 29;35(6):1727-1751. doi: 10.1093/plcell/koad049. Plant Cell. 2023. PMID: 36807982 Free PMC article. Review.
References
-
- Aigner H, Wilson RH, Bracher A, Calisse L, Bhat JY, Hartl FU, Hayer-Hartl M (2017) Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2. Science 358: 1272–1278 - PubMed
-
- Andersson I, Backlund A (2008) Structure and function of Rubisco. Plant Physiol Biochem 46: 275–291 - PubMed
-
- Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65: 415–442 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

