Bioinformatics Analysis of SARS-CoV-2 to Approach an Effective Vaccine Candidate Against COVID-19
- PMID: 33625681
- PMCID: PMC7902242
- DOI: 10.1007/s12033-021-00303-0
Bioinformatics Analysis of SARS-CoV-2 to Approach an Effective Vaccine Candidate Against COVID-19
Abstract
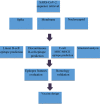
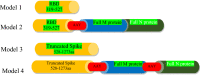
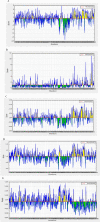
The emerging Coronavirus Disease 2019 (COVID-19) pandemic has posed a serious threat to the public health worldwide, demanding urgent vaccine provide. According to the virus feature as an RNA virus, a high rate of mutations imposes some vaccine design difficulties. Bioinformatics tools have been widely used to make advantage of conserved regions as well as immunogenicity. In this study, we aimed at immunoinformatic evaluation of SARS-CoV-2 proteins conservancy and immunogenicity to design a preventive vaccine candidate. Spike, Membrane and Nucleocapsid amino acid sequences were obtained, and four possible fusion proteins were assessed and compared in terms of structural features and immunogenicity, and population coverage. MHC-I and MHC-II T-cell epitopes, the linear and conformational B-cell epitopes were evaluated. Among the predicted models, the truncated form of Spike in fusion with M and N protein applying AAY linker has high rate of MHC-I and MCH-II epitopes with high antigenicity and acceptable population coverage of 82.95% in Iran and 92.51% in Europe. The in silico study provided truncated Spike-M-N SARS-CoV-2 as a potential preventive vaccine candidate for further in vivo evaluation.
Keywords: COVID-19; In silico; Protein; SARS-CoV-2; Vaccine design.
Conflict of interest statement
The authors declare no potential conflict of interest that could negatively influence the study.
Figures

References
-
- World Health Organization . WHO Coronavirus Disease (COVID-19) Dashboard. Geneva: World Health Organization; 2020.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

