Ultra-rapid detection of SARS-CoV-2 in public workspace environments
- PMID: 33626039
- PMCID: PMC7904170
- DOI: 10.1371/journal.pone.0240524
Ultra-rapid detection of SARS-CoV-2 in public workspace environments
Abstract
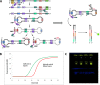
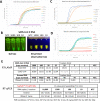
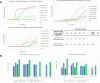
Managing the pandemic caused by SARS-CoV-2 requires new capabilities in testing, including the possibility of identifying, in minutes, infected individuals as they enter spaces where they must congregate in a functioning society, including workspaces, schools, points of entry, and commercial business establishments. Here, the only useful tests (a) require no sample transport, (b) require minimal sample manipulation, (c) can be performed by unlicensed individuals, (d) return results on the spot in much less than one hour, and (e) cost no more than a few dollars. The sensitivity need not be as high as normally required by the FDA for screening asymptomatic carriers (as few as 10 virions per sample), as these viral loads are almost certainly not high enough for an individual to present a risk for forward infection. This allows tests specifically useful for this pandemic to trade-off unneeded sensitivity for necessary speed, simplicity, and frugality. In some studies, it was shown that viral load that creates forward-infection risk may exceed 105 virions per milliliter, easily within the sensitivity of an RNA amplification architecture, but unattainable by antibody-based architectures that simply target viral antigens. Here, we describe such a test based on a displaceable probe loop amplification architecture.
Conflict of interest statement
Firebird Biomolecular Sciences, LLC, GenePath Diagnostics, Inc., and GenePath Diagnostics India Pvt. Ltd. employ the indicated authors and the specific roles of these authors are articulated in the ‘author contributions’ section. OY, ZY, SAB and their institutions own intellectual property associated with the assay. Some of the items mentioned here are sold by Firebird Biomolecular Sciences, LLC, which employs the indicated authors and is owned by SAB. This does not alter the authors’ and institutions’ adherence to PLOS ONE policies on sharing data and materials.
Figures


Similar articles
-
Saliva TwoStep for rapid detection of asymptomatic SARS-CoV-2 carriers.Elife. 2021 Mar 29;10:e65113. doi: 10.7554/eLife.65113. Elife. 2021. PMID: 33779548 Free PMC article.
-
Effectiveness and cost-effectiveness of four different strategies for SARS-CoV-2 surveillance in the general population (CoV-Surv Study): a structured summary of a study protocol for a cluster-randomised, two-factorial controlled trial.Trials. 2021 Jan 8;22(1):39. doi: 10.1186/s13063-020-04982-z. Trials. 2021. PMID: 33419461 Free PMC article.
-
Rapid diagnostics for SARS-CoV-2 virus: point-of-care testing and lessons learned during the pandemic.Bioanalysis. 2021 Aug;13(15):1165-1167. doi: 10.4155/bio-2021-0100. Epub 2021 Jul 21. Bioanalysis. 2021. PMID: 34286599 Free PMC article. No abstract available.
-
Isothermal SARS-CoV-2 Diagnostics: Tools for Enabling Distributed Pandemic Testing as a Means of Supporting Safe Reopenings.ACS Synth Biol. 2020 Nov 20;9(11):2861-2880. doi: 10.1021/acssynbio.0c00359. Epub 2020 Oct 9. ACS Synth Biol. 2020. PMID: 32966744 Review.
-
SARS-CoV-2 and Variant Diagnostic Testing Approaches in the United States.Viruses. 2021 Dec 13;13(12):2492. doi: 10.3390/v13122492. Viruses. 2021. PMID: 34960762 Free PMC article. Review.
Cited by
-
A LAMP sequencing approach for high-throughput co-detection of SARS-CoV-2 and influenza virus in human saliva.Elife. 2022 May 9;11:e69949. doi: 10.7554/eLife.69949. Elife. 2022. PMID: 35532013 Free PMC article.
-
Development of an Integrated Sample Amplification Control for Salivary Point-of-Care Pathogen Testing.medRxiv [Preprint]. 2023 Oct 3:2023.10.03.23296477. doi: 10.1101/2023.10.03.23296477. medRxiv. 2023. Update in: Anal Chim Acta. 2024 Jan 25;1287:342072. doi: 10.1016/j.aca.2023.342072. PMID: 37873363 Free PMC article. Updated. Preprint.
-
Development of an integrated sample amplification control for salivary point-of-care pathogen testing.Anal Chim Acta. 2024 Jan 25;1287:342072. doi: 10.1016/j.aca.2023.342072. Epub 2023 Nov 29. Anal Chim Acta. 2024. PMID: 38182338 Free PMC article.
-
Development of a Duplex LAMP Assay with Probe-Based Readout for Simultaneous Real-Time Detection of Schistosoma mansoni and Strongyloides spp. -A Laboratory Approach to Point-Of-Care.Int J Mol Sci. 2023 Jan 3;24(1):893. doi: 10.3390/ijms24010893. Int J Mol Sci. 2023. PMID: 36614336 Free PMC article.
-
Accessible LAMP-Enabled Rapid Test (ALERT) for Detecting SARS-CoV-2.Viruses. 2021 Apr 23;13(5):742. doi: 10.3390/v13050742. Viruses. 2021. PMID: 33922716 Free PMC article.
References
-
- Riou J, Althaus CL. Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. Euro surveillance: bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2020;25(4):2000058 10.2807/1560-7917.ES.2020.25.4.2000058 . - DOI - PMC - PubMed
-
- Benner SA, Gaucher EA, Li T. Post-genomic evolutionary analyses of the severe acute respiratory syndrome (SARS) virus genome using the Master Catalog[R] interpretive proteomics platform. PharmaGenomics. 2003;3:S17.
-
- Chan JF, Lau SK, To KK, Cheng VC, Woo PC, Yuen KY. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev 2015;28(1098–6618 (Electronic)):465–522. doi: D—NLM: PMC4402954 [Available on 04/01/16] EDAT- 2015/03/27 06:00 MHDA- 2015/04/22 06:00 CRDT- 2015/03/27 06:00 PMCR- 2016/04/01 00:00 AID—28/2/465 [pii] AID—10.1128/CMR.00102-14 PST—ppublish. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

