Dedicator of Cytokinesis 5 Regulates Keratinocyte Function and Promotes Diabetic Wound Healing
- PMID: 33627322
- PMCID: PMC8173801
- DOI: 10.2337/db20-1008
Dedicator of Cytokinesis 5 Regulates Keratinocyte Function and Promotes Diabetic Wound Healing
Abstract
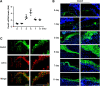
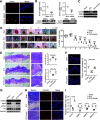
Cutaneous wound healing is a fundamental biologic and coordinated process, and failure to maintain this process contributes to the dysfunction of tissue homeostasis, increasing the global burden of diabetic foot ulcerations. However, the factors that mediate this process are not fully understood. Here, we identify the pivotal role of dedicator of cytokinesis 5 (Dock5) in keratinocyte functions contributing to the process of skin wound healing. Specifically, Dock5 is highly upregulated during the proliferative phase of wound repair and is predominantly expressed in epidermal keratinocytes. It regulates keratinocyte adhesion, migration, and proliferation and influences the functions of extracellular matrix (ECM) deposition by facilitating the ubiquitination of transcription factor ZEB1 to activate laminin-332/integrin signaling. Genetic ablation of Dock5 in mice leads to attenuated reepithelialization and granulation tissue formation, and Dock5 overexpression-improved skin repair can be abrogated by LAMA3 knockdown. Importantly, Dock5 expression in the skin edge is reduced in patients and animal models of diabetes, further suggesting a direct correlation between its abundance and healing capability. The rescue of Dock5 expression in diabetic mice causes a significant improvement in reepithelialization, collagen deposition, ECM production, and granulation. Our study provides a potential therapeutic target for wound healing impairment during diabetes.
© 2021 by the American Diabetes Association.
Figures




Similar articles
-
Liraglutide Promotes Diabetic Wound Healing via Myo1c/Dock5.Adv Sci (Weinh). 2024 Oct;11(39):e2405987. doi: 10.1002/advs.202405987. Epub 2024 Aug 19. Adv Sci (Weinh). 2024. PMID: 39159301 Free PMC article.
-
Interleukin-22 Promotes Wound Repair in Diabetes by Improving Keratinocyte Pro-Healing Functions.J Invest Dermatol. 2015 Nov;135(11):2862-2870. doi: 10.1038/jid.2015.278. Epub 2015 Jul 13. J Invest Dermatol. 2015. PMID: 26168231
-
Myostatin-null mice exhibit delayed skin wound healing through the blockade of transforming growth factor-β signaling by decorin.Am J Physiol Cell Physiol. 2012 Apr 15;302(8):C1213-25. doi: 10.1152/ajpcell.00179.2011. Epub 2012 Jan 25. Am J Physiol Cell Physiol. 2012. PMID: 22277753
-
The role of keratinocyte function on the defected diabetic wound healing.Int J Burns Trauma. 2021 Dec 15;11(6):430-441. eCollection 2021. Int J Burns Trauma. 2021. PMID: 35111377 Free PMC article. Review.
-
Focal Contact and Hemidesmosomal Proteins in Keratinocyte Migration and Wound Repair.Adv Wound Care (New Rochelle). 2014 Mar 1;3(3):247-263. doi: 10.1089/wound.2013.0489. Adv Wound Care (New Rochelle). 2014. PMID: 24669360 Free PMC article. Review.
Cited by
-
Mitochondrial glycerol 3-phosphate dehydrogenase deficiency exacerbates lipotoxic cardiomyopathy.iScience. 2024 May 11;27(6):109796. doi: 10.1016/j.isci.2024.109796. eCollection 2024 Jun 21. iScience. 2024. PMID: 38832016 Free PMC article.
-
The mouse resource at National Resource Center for Mutant Mice.Mamm Genome. 2022 Mar;33(1):143-156. doi: 10.1007/s00335-021-09940-x. Epub 2022 Feb 9. Mamm Genome. 2022. PMID: 35138443 Review.
-
Dock5 activation facilitates diabetic wound healing.Metabol Open. 2021 Mar 17;10:100088. doi: 10.1016/j.metop.2021.100088. eCollection 2021 Jun. Metabol Open. 2021. PMID: 33855290 Free PMC article. No abstract available.
-
Human Keratinocyte-Derived Exosomal MALAT1 Promotes Diabetic Wound Healing by Upregulating MFGE8 via microRNA-1914-3p.Int J Nanomedicine. 2023 Feb 21;18:949-970. doi: 10.2147/IJN.S399785. eCollection 2023. Int J Nanomedicine. 2023. PMID: 36852184 Free PMC article.
-
Platelet-Rich Plasma-Derived Exosomal USP15 Promotes Cutaneous Wound Healing via Deubiquitinating EIF4A1.Oxid Med Cell Longev. 2021 Aug 9;2021:9674809. doi: 10.1155/2021/9674809. eCollection 2021. Oxid Med Cell Longev. 2021. Retraction in: Oxid Med Cell Longev. 2024 Jan 9;2024:9808072. doi: 10.1155/2024/9808072. PMID: 34422211 Free PMC article. Retracted.
References
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

