Loss of Optineurin Drives Cancer Immune Evasion via Palmitoylation-Dependent IFNGR1 Lysosomal Sorting and Degradation
- PMID: 33627378
- PMCID: PMC8292167
- DOI: 10.1158/2159-8290.CD-20-1571
Loss of Optineurin Drives Cancer Immune Evasion via Palmitoylation-Dependent IFNGR1 Lysosomal Sorting and Degradation
Abstract
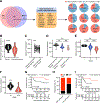
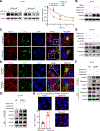
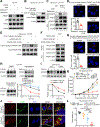
Mutations in IFN and MHC signaling genes endow immunotherapy resistance. Patients with colorectal cancer infrequently exhibit IFN and MHC signaling gene mutations and are generally resistant to immunotherapy. In exploring the integrity of IFN and MHC signaling in colorectal cancer, we found that optineurin was a shared node between the two pathways and predicted colorectal cancer patient outcome. Loss of optineurin occurs in early-stage human colorectal cancer. Immunologically, optineurin deficiency was shown to attenuate IFNGR1 and MHC-I expression, impair T-cell immunity, and diminish immunotherapy efficacy in murine cancer models and patients with cancer. Mechanistically, we observed that IFNGR1 was S-palmitoylated on Cys122, and AP3D1 bound with and sorted palmitoylated IFNGR1 to lysosome for degradation. Unexpectedly, optineurin interacted with AP3D1 to prevent palmitoylated IFNGR1 lysosomal sorting and degradation, thereby maintaining IFNγ and MHC-I signaling integrity. Furthermore, pharmacologically targeting IFNGR1 palmitoylation stabilized IFNGR1, augmented tumor immunity, and sensitized checkpoint therapy. Thus, loss of optineurin drives immune evasion and intrinsic immunotherapy resistance in colorectal cancer. SIGNIFICANCE: Loss of optineurin impairs the integrity of both IFNγ and MHC-I signaling pathways via palmitoylation-dependent IFNGR1 lysosomal sorting and degradation, thereby driving immune evasion and intrinsic immunotherapy resistance in colorectal cancer. Our work suggests that pharmacologically targeting IFNGR1 palmitoylation can stabilize IFNGR1, enhance T-cell immunity, and sensitize checkpoint therapy in colorectal cancer.See related commentary by Salvagno and Cubillos-Ruiz, p. 1623.This article is highlighted in the In This Issue feature, p. 1601.
©2021 American Association for Cancer Research.
Conflict of interest statement
Figures
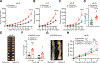


Comment in
-
Optineurin Guards IFNγ Signaling in Cancer Cells.Cancer Discov. 2021 Jul;11(7):1623-1625. doi: 10.1158/2159-8290.CD-21-0362. Cancer Discov. 2021. PMID: 34284996
References
-
- Gryfe R, Kim H, Hsieh ET, Aronson MD, Holowaty EJ, Bull SB, et al. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med 2000;342(2):69–77. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

