Primer, Pipelines, Parameters: Issues in 16S rRNA Gene Sequencing
- PMID: 33627512
- PMCID: PMC8544895
- DOI: 10.1128/mSphere.01202-20
Primer, Pipelines, Parameters: Issues in 16S rRNA Gene Sequencing
Abstract
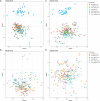
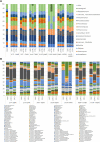
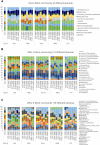
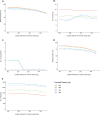
Short-amplicon 16S rRNA gene sequencing is currently the method of choice for studies investigating microbiomes. However, comparative studies on differences in procedures are scarce. We sequenced human stool samples and mock communities with increasing complexity using a variety of commonly used protocols. Short amplicons targeting different variable regions (V-regions) or ranges thereof (V1-V2, V1-V3, V3-V4, V4, V4-V5, V6-V8, and V7-V9) were investigated for differences in the composition outcome due to primer choices. Next, the influence of clustering (operational taxonomic units [OTUs], zero-radius OTUs [zOTUs], and amplicon sequence variants [ASVs]), different databases (GreenGenes, the Ribosomal Database Project, Silva, the genomic-based 16S rRNA Database, and The All-Species Living Tree), and bioinformatic settings on taxonomic assignment were also investigated. We present a systematic comparison across all typically used V-regions using well-established primers. While it is known that the primer choice has a significant influence on the resulting microbial composition, we show that microbial profiles generated using different primer pairs need independent validation of performance. Further, comparing data sets across V-regions using different databases might be misleading due to differences in nomenclature (e.g., Enterorhabdus versus Adlercreutzia) and varying precisions in classification down to genus level. Overall, specific but important taxa are not picked up by certain primer pairs (e.g., Bacteroidetes is missed using primers 515F-944R) or due to the database used (e.g., Acetatifactor in GreenGenes and the genomic-based 16S rRNA Database). We found that appropriate truncation of amplicons is essential and different truncated-length combinations should be tested for each study. Finally, specific mock communities of sufficient and adequate complexity are highly recommended.IMPORTANCE In 16S rRNA gene sequencing, certain bacterial genera were found to be underrepresented or even missing in taxonomic profiles when using unsuitable primer combinations, outdated reference databases, or inadequate pipeline settings. Concerning the last, quality thresholds as well as bioinformatic settings (i.e., clustering approach, analysis pipeline, and specific adjustments such as truncation) are responsible for a number of observed differences between studies. Conclusions drawn by comparing one data set to another (e.g., between publications) appear to be problematic and require independent cross-validation using matching V-regions and uniform data processing. Therefore, we highlight the importance of a thought-out study design including sufficiently complex mock standards and appropriate V-region choice for the sample of interest. The use of processing pipelines and parameters must be tested beforehand.
Keywords: 16S rRNA gene sequencing; amplicon sequencing; bioinformatic settings; clustering; databases; microbiome; mock communities; variable regions.
Copyright © 2021 Abellan-Schneyder et al.
Figures


References
-
- Reitmeier S, Kiessling S, Clavel T, List M, Almeida EL, Ghosh TS, Neuhaus K, Grallert H, Linseisen J, Skurk T, Brandl B, Breuninger TA, Troll M, Rathmann W, Linkohr B, Hauner H, Laudes M, Franke A, Le Roy CI, Bell JT, Spector T, Baumbach J, O’Toole PW, Peters A, Haller D. 2020. Arrhythmic gut microbiome signatures predict risk of type 2 diabetes. Cell Host Microbe 28:258–272.e6. doi:10.1016/j.chom.2020.06.004. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

