Aging and rejuvenation - a modular epigenome model
- PMID: 33627519
- PMCID: PMC7950254
- DOI: 10.18632/aging.202712
Aging and rejuvenation - a modular epigenome model
Abstract
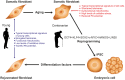
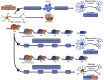
The view of aging has evolved in parallel with the advances in biomedical sciences. Long considered as an irreversible process where interventions were only aimed at slowing down its progression, breakthrough discoveries like animal cloning and cell reprogramming have deeply changed our understanding of postnatal development, giving rise to the emerging view that the epigenome is the driver of aging. The idea was significantly strengthened by the converging discovery that DNA methylation (DNAm) at specific CpG sites could be used as a highly accurate biomarker of age defined by an algorithm known as the Horvath clock. It was at this point where epigenetic rejuvenation came into play as a strategy to reveal to what extent biological age can be set back by making the clock tick backwards. Initial evidence suggests that when the clock is forced to tick backwards in vivo, it is only able to drag the phenotype to a partially rejuvenated condition. In order to explain the results, a bimodular epigenome is proposed, where module A represents the DNAm clock component and module B the remainder of the epigenome. Epigenetic rejuvenation seems to hold the key to arresting or even reversing organismal aging.
Keywords: DNA methylation; aging; cell reprogramming; epigenetic clock; rejuvenation.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

