Comparison of rhesus and cynomolgus macaques as an infection model for COVID-19
- PMID: 33627662
- PMCID: PMC7904795
- DOI: 10.1038/s41467-021-21389-9
Comparison of rhesus and cynomolgus macaques as an infection model for COVID-19
Abstract
A novel coronavirus, SARS-CoV-2, has been identified as the causative agent of the current COVID-19 pandemic. Animal models, and in particular non-human primates, are essential to understand the pathogenesis of emerging diseases and to assess the safety and efficacy of novel vaccines and therapeutics. Here, we show that SARS-CoV-2 replicates in the upper and lower respiratory tract and causes pulmonary lesions in both rhesus and cynomolgus macaques. Immune responses against SARS-CoV-2 are also similar in both species and equivalent to those reported in milder infections and convalescent human patients. This finding is reiterated by our transcriptional analysis of respiratory samples revealing the global response to infection. We describe a new method for lung histopathology scoring that will provide a metric to enable clearer decision making for this key endpoint. In contrast to prior publications, in which rhesus are accepted to be the preferred study species, we provide convincing evidence that both macaque species authentically represent mild to moderate forms of COVID-19 observed in the majority of the human population and both species should be used to evaluate the safety and efficacy of interventions against SARS-CoV-2. Importantly, accessing cynomolgus macaques will greatly alleviate the pressures on current rhesus stocks.
Conflict of interest statement
The authors declare no competing interests.
Figures


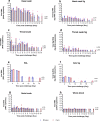


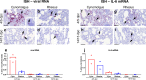
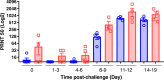
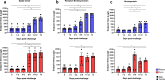

References
-
- WHO. Coronavirus disease (COVID-19) Weekly Epidemiological Update and Weekly Operational Update. 24 November 2020. https://www.who.int/publications/m/item/weekly-epidemiological-update---....
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

