Use of stable isotope-tagged thymidine and multi-isotope imaging mass spectrometry (MIMS) for quantification of human cardiomyocyte division
- PMID: 33627842
- PMCID: PMC8221415
- DOI: 10.1038/s41596-020-00477-y
Use of stable isotope-tagged thymidine and multi-isotope imaging mass spectrometry (MIMS) for quantification of human cardiomyocyte division
Abstract
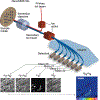
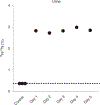
Quantification of cellular proliferation in humans is important for understanding biology and responses to injury and disease. However, existing methods require administration of tracers that cannot be ethically administered in humans. We present a protocol for the direct quantification of cellular proliferation in human hearts. The protocol involves administration of non-radioactive, non-toxic stable isotope 15Nitrogen-enriched thymidine (15N-thymidine), which is incorporated into DNA during S-phase, in infants with tetralogy of Fallot, a common form of congenital heart disease. Infants with tetralogy of Fallot undergo surgical repair, which requires the removal of pieces of myocardium that would otherwise be discarded. This protocol allows for the quantification of cardiomyocyte proliferation in this discarded tissue. We quantitatively analyzed the incorporation of 15N-thymidine with multi-isotope imaging spectrometry (MIMS) at a sub-nuclear resolution, which we combined with correlative confocal microscopy to quantify formation of binucleated cardiomyocytes and cardiomyocytes with polyploid nuclei. The entire protocol spans 3-8 months, which is dependent on the timing of surgical repair, and 3-4.5 researcher days. This protocol could be adapted to study cellular proliferation in a variety of human tissues.
Figures
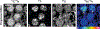





References
-
- Romar GA, Kupper TS & Divito SJ Research techniques made simple: techniques to assess cell proliferation. J. Invest. Dermatol 136, e1–e7 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

