Evaluation of full-length nanopore 16S sequencing for detection of pathogens in microbial keratitis
- PMID: 33628638
- PMCID: PMC7891086
- DOI: 10.7717/peerj.10778
Evaluation of full-length nanopore 16S sequencing for detection of pathogens in microbial keratitis
Abstract
Background: Microbial keratitis is a leading cause of preventable blindness worldwide. Conventional sampling and culture techniques are time-consuming, with over 40% of cases being culture-negative. Nanopore sequencing technology is portable and capable of generating long sequencing reads in real-time. The aim of this study is to evaluate the potential of nanopore sequencing directly from clinical samples for the diagnosis of bacterial microbial keratitis.
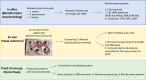
Methods: Using full-length 16S rRNA amplicon sequences from a defined mock microbial community, we evaluated and benchmarked our bioinformatics analysis pipeline for taxonomic assignment on three different 16S rRNA databases (NCBI 16S RefSeq, RDP and SILVA) with clustering at 97%, 99% and 100% similarities. Next, we optimised the sample collection using an ex vivo porcine model of microbial keratitis to compare DNA recovery rates of 12 different collection methods: 21-gauge needle, PTFE membrane (4 mm and 6 mm), Isohelix™ SK-2S, Sugi® Eyespear, Cotton, Rayon, Dryswab™, Hydraflock®, Albumin-coated, Purflock®, Purfoam and Polyester swabs. As a proof-of-concept study, we then used the sampling technique that provided the highest DNA recovery, along with the optimised bioinformatics pipeline, to prospectively collected samples from patients with suspected microbial keratitis. The resulting nanopore sequencing results were then compared to standard microbiology culture methods.
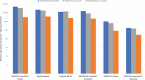
Results: We found that applying alignment filtering to nanopore sequencing reads and aligning to the NCBI 16S RefSeq database at 100% similarity provided the most accurate bacterial taxa assignment. DNA concentration recovery rates differed significantly between the collection methods (p < 0.001), with the Sugi® Eyespear swab providing the highest mean rank of DNA concentration. Then, applying the optimised collection method and bioinformatics pipeline directly to samples from two patients with suspected microbial keratitis, sequencing results from Patient A were in agreement with culture results, whilst Patient B, with negative culture results and previous antibiotic use, showed agreement between nanopore and Illumina Miseq sequencing results.
Conclusion: We have optimised collection methods and demonstrated a novel workflow for identification of bacterial microbial keratitis using full-length 16S nanopore sequencing.
Keywords: 16S bioinformatics; Cornea infection; Corneal infection; Eye infection; Eye swab; Full length 16S rRNA sequencing; Microbial keratitis; Molecular diagnostics; Nanopore sequencing; Ophthalmology.
© 2021 Low et al.
Conflict of interest statement
Professor Nicholas J. Loman is an Academic Editor for PeerJ and has received Oxford Nanopore Technologies (ONT) reagents free of charge to support his research programme (but not for this study), travel expenses to speak at ONT events and an honorarium to speak at an ONT company meeting. All the swabs and collection methods were provided free of charge directly from the suppliers. Nicholas J. Loman is a director of Microbial Genomics Ltd. Pablo Fuentes-Utrilla is an employee of Microbial Genomics Ltd.
Figures
References
-
- 16S Sequencing and Analysis 16S analysis using real-time, long-read nanopore sequencing. 2020. https://nanoporetech.com/analyse/16s. [21 July 2020]. https://nanoporetech.com/analyse/16s
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

