Construction and Analysis of Survival-Associated Competing Endogenous RNA Network in Lung Adenocarcinoma
- PMID: 33628780
- PMCID: PMC7895565
- DOI: 10.1155/2021/4093426
Construction and Analysis of Survival-Associated Competing Endogenous RNA Network in Lung Adenocarcinoma
Abstract
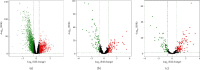
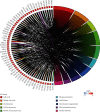
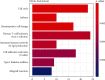
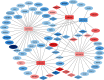
Increasing evidence has shown that noncoding RNAs play significant roles in the initiation, progression, and metastasis of tumours via participating in competing endogenous RNA (ceRNA) networks. However, the survival-associated ceRNA in lung adenocarcinoma (LUAD) remains poorly understood. In this study, we aimed to investigate the regulatory mechanisms underlying ceRNA in LUAD to identify novel prognostic factors. mRNA, lncRNA, and miRNA sequencing data obtained from the GDC data portal were utilized to identify differentially expressed (DE) RNAs. Survival-related RNAs were recognized using univariate Kaplan-Meier survival analysis. We performed functional enrichment analysis of survival-related mRNAs using the clusterProfiler package of R and STRING. lncRNA-miRNA and miRNA-mRNA interactions were predicted based on miRcode, Starbase, and miRanda. Subsequently, the survival-associated ceRNA network was constructed for LUAD. Multivariate Cox regression analysis was used to identify prognostic factors. Finally, we acquired 15 DE miRNAs, 49 DE lncRNAs, and 843 DE mRNAs associated with significant overall survival. Functional enrichment analysis indicated that survival-related DE mRNAs were enriched in cell cycle. The survival-associated lncRNA-miRNA-mRNA ceRNA network was constructed using five miRNAs, 49 mRNAs, and 21 lncRNAs. Furthermore, seven hub RNAs (LINC01936, miR-20a-5p, miR-31-5p, TNS1, TGFBR2, SMAD7, and NEDD4L) were identified based on the ceRNA network. LINC01936 and miR-31-5p were found to be significant using the multifactorial Cox regression model. In conclusion, we successfully constructed a survival-related lncRNA-miRNA-mRNA ceRNA regulatory network in LUAD and identified seven hub RNAs, which provide novel insights into the regulatory molecular mechanisms associated with survival of LUAD, and identified two independent prognostic predictors for LUAD.
Copyright © 2021 Lixian Chen et al.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this paper.
Figures





Similar articles
-
Predictions of the dysregulated competing endogenous RNA signature involved in the progression of human lung adenocarcinoma.Cancer Biomark. 2020;29(3):399-416. doi: 10.3233/CBM-200133. Cancer Biomark. 2020. PMID: 32741804
-
Characterization of a non-coding RNA-associated ceRNA network in metastatic lung adenocarcinoma.J Cell Mol Med. 2020 Oct;24(20):11680-11690. doi: 10.1111/jcmm.15778. Epub 2020 Aug 29. J Cell Mol Med. 2020. PMID: 32860342 Free PMC article.
-
Integrative Analysis of Three Novel Competing Endogenous RNA Biomarkers with a Prognostic Value in Lung Adenocarcinoma.Biomed Res Int. 2020 Aug 4;2020:2837906. doi: 10.1155/2020/2837906. eCollection 2020. Biomed Res Int. 2020. PMID: 32802839 Free PMC article.
-
Reconstruction and Analysis of the Differentially Expressed IncRNA-miRNA-mRNA Network Based on Competitive Endogenous RNA in Hepatocellular Carcinoma.Crit Rev Eukaryot Gene Expr. 2019;29(6):539-549. doi: 10.1615/CritRevEukaryotGeneExpr.2019028740. Crit Rev Eukaryot Gene Expr. 2019. PMID: 32422009 Review.
-
Bioinformatics analysis of ceRNA network of autophagy-related genes in pediatric asthma.Medicine (Baltimore). 2023 Dec 1;102(48):e36343. doi: 10.1097/MD.0000000000036343. Medicine (Baltimore). 2023. PMID: 38050261 Free PMC article. Review.
Cited by
-
Six MicroRNA Prognostic Models for Overall Survival of Lung Adenocarcinoma.Genet Res (Camb). 2022 Aug 27;2022:5955052. doi: 10.1155/2022/5955052. eCollection 2022. Genet Res (Camb). 2022. PMID: 36101742 Free PMC article.
-
LINC01936 inhibits the proliferation and metastasis of lung squamous cell carcinoma probably by EMT signaling and immune infiltration.PeerJ. 2023 Dec 7;11:e16447. doi: 10.7717/peerj.16447. eCollection 2023. PeerJ. 2023. PMID: 38084139 Free PMC article.
-
TNS1: Emerging Insights into Its Domain Function, Biological Roles, and Tumors.Biology (Basel). 2022 Oct 26;11(11):1571. doi: 10.3390/biology11111571. Biology (Basel). 2022. PMID: 36358270 Free PMC article. Review.
-
Necroptosis-Related LncRNA Signatures for Prognostic Prediction in Uterine Corpora Endometrial Cancer.Reprod Sci. 2023 Feb;30(2):576-589. doi: 10.1007/s43032-022-01023-9. Epub 2022 Jul 19. Reprod Sci. 2023. PMID: 35854199 Free PMC article.
-
Identification and Construction of a Predictive Immune-Related lncRNA Signature Model for Melanoma.Int J Gen Med. 2021 Dec 1;14:9227-9235. doi: 10.2147/IJGM.S340025. eCollection 2021. Int J Gen Med. 2021. PMID: 34880662 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

