Genomic Expression Profiling and Bioinformatics Analysis of Chronic Recurrent Multifocal Osteomyelitis
- PMID: 33628812
- PMCID: PMC7888306
- DOI: 10.1155/2021/6898093
Genomic Expression Profiling and Bioinformatics Analysis of Chronic Recurrent Multifocal Osteomyelitis
Abstract
Objective: Chronic nonbacterial osteomyelitis (CNO) is an autoinflammatory bone disorder. Its most severe form is referred to as chronic recurrent multifocal osteomyelitis (CRMO). Currently, the exact molecular pathophysiology of CNO/CRMO remains unknown. No uniform diagnostic standard and treatment protocol were available for this disease. The aim of this study was to identify the differentially expressed genes (DEGs) in CRMO tissues compared to normal control tissues to investigate the mechanisms of CRMO.
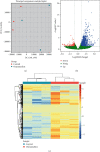
Materials: Microarray data from the GSE133378 (12 CRMO and 148 matched normal tissue samples) data sets were downloaded from the Gene Expression Omnibus (GEO) database. DEGs were identified using the limma package in the R software. Gene Ontology (GO) analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, and protein-protein interaction (PPI) network analysis were performed to further investigate the function of the identified DEGs.
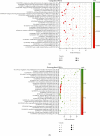
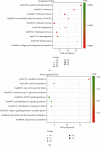
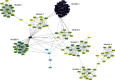
Results: This study identified a total of 1299 differentially expressed mRNAs, including1177 upregulated genes and 122 downregulated genes, between CRMO and matched normal tissue samples. GO analyses showed that DEGs were enriched in immune-related terms. KEGG pathway enrichment analyses showed that the DEGs were mainly related to oxidative phosphorylation, ribosome, and Parkinson disease. Eight modules were extracted from the gene expression network, including one module constituted with immune-related genes and one module constituted with ribosomal-related genes.
Conclusion: Oxidative phosphorylation, ribosome, and Parkinson disease pathways were significantly associated with CRMO. The immune-related genes including IRF5, OAS3, and HLA-A, as well as numerous ribosomal-related genes, might be implicated in the pathogenesis of CRMO. The identification of these genes may contribute to the development of early diagnostic tools, prognostic markers, or therapeutic targets in CRMO.
Copyright © 2021 Kai Huang et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures
Similar articles
-
Integrated bioinformatics analysis for differentially expressed genes and signaling pathways identification in gastric cancer.Int J Med Sci. 2021 Jan 1;18(3):792-800. doi: 10.7150/ijms.47339. eCollection 2021. Int J Med Sci. 2021. PMID: 33437215 Free PMC article.
-
Bioinformatics analyses of significant genes, related pathways and candidate prognostic biomarkers in glioblastoma.Mol Med Rep. 2018 Nov;18(5):4185-4196. doi: 10.3892/mmr.2018.9411. Epub 2018 Aug 21. Mol Med Rep. 2018. PMID: 30132538 Free PMC article.
-
Bioinformatics analysis of gene expression profile data to screen key genes involved in intracranial aneurysms.Mol Med Rep. 2019 Nov;20(5):4415-4424. doi: 10.3892/mmr.2019.10696. Epub 2019 Sep 23. Mol Med Rep. 2019. PMID: 31545495 Free PMC article.
-
Chronic Recurrent Multifocal Osteomyelitis (CRMO): Presentation, Pathogenesis, and Treatment.Curr Osteoporos Rep. 2017 Dec;15(6):542-554. doi: 10.1007/s11914-017-0405-9. Curr Osteoporos Rep. 2017. PMID: 29080202 Free PMC article. Review.
-
Chronic Recurrent Multifocal Osteomyelitis (CRMO): A Study of 12 Cases from One Institution and Literature Review.Fetal Pediatr Pathol. 2022 Oct;41(5):759-770. doi: 10.1080/15513815.2021.1978602. Epub 2021 Sep 20. Fetal Pediatr Pathol. 2022. PMID: 34542007 Review.
Cited by
-
Pyoderma gangrenosum following anti-TNF therapy in chronic recurrent multifocal osteomyelitis: drug reaction or cutaneous manifestation of the disease? A critical review on the topic with an emblematic case report.Front Med (Lausanne). 2023 May 31;10:1197273. doi: 10.3389/fmed.2023.1197273. eCollection 2023. Front Med (Lausanne). 2023. PMID: 37324147 Free PMC article. Review.
-
Screening and identification of osteoarthritis related differential genes and construction of a risk prognosis model based on bioinformatics analysis.Ann Transl Med. 2022 Apr;10(8):444. doi: 10.21037/atm-22-1135. Ann Transl Med. 2022. PMID: 35571384 Free PMC article.
-
A Successful Bisphosphonates Monotherapy in Spinal Form of Paediatric Chronic Recurrent Multifocal Osteomyelitis (CRMO)-Case Report.Metabolites. 2023 Feb 25;13(3):344. doi: 10.3390/metabo13030344. Metabolites. 2023. PMID: 36984784 Free PMC article.
-
Sterile osteomyelitis: a cardinal sign of autoinflammation.Reumatologia. 2024;62(6):475-488. doi: 10.5114/reum/196595. Epub 2024 Dec 24. Reumatologia. 2024. PMID: 39866303 Free PMC article. Review.
References
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

