Identification of a New Prognostic Risk Signature of Clear Cell Renal Cell Carcinoma Based on N6-Methyladenosine RNA Methylation Regulators
- PMID: 33628845
- PMCID: PMC7895564
- DOI: 10.1155/2021/6617841
Identification of a New Prognostic Risk Signature of Clear Cell Renal Cell Carcinoma Based on N6-Methyladenosine RNA Methylation Regulators
Abstract
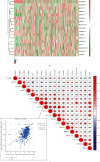
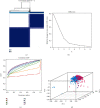
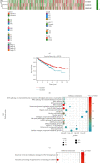
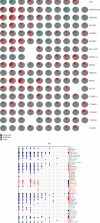
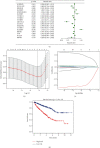
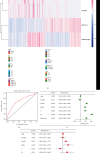
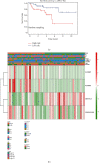
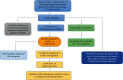
As the most prevalent internal eukaryotic modification, N6-methyladenosine (m6A) is installed by methyltransferases, removed by demethylases, and recognized by readers. However, there are few studies on the role of m6A in clear cell renal cell carcinoma (ccRCC). In this study, we researched the RNA-seq transcriptome data of ccRCC in the TCGA dataset and used bioinformatics analyses to detect the relationship between m6A RNA methylation regulators and ccRCC. First, we compared the expression of 18 m6A RNA methylation regulators in ccRCC patients and normal tissues. Then, data from ccRCC patients were divided into two clusters by consensus clustering. LASSO Cox regression analysis was used to build a risk signature to predict the prognosis of patients with ccRCC. An ROC curve, univariate Cox regression analysis, and multivariate Cox regression analysis were used to verify this risk signature's predictive ability. Then, we internally validated this signature by random sampling. Finally, we explored the role of the genes in the signature in some common pathways. Gene distribution between the two subgroups was different; cluster 2 was gender-related and had a worse prognosis. IGF2BP3, IGF2BP2, HNRNPA2B1, and METTL14 were chosen to build the risk signature. The overall survival of the high- and low-risk groups was significantly different (p = 7.47e - 12). The ROC curve also indicated that the risk signature had a decent predictive significance (AUC = 0.72). These results imply that the risk signature has a potential value for ccRCC treatment.
Copyright © 2021 Yan Zhang et al.
Conflict of interest statement
The authors declare that there is no conflict of interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

