A Biochemical Perspective of the Nonstructural Proteins (NSPs) and the Spike Protein of SARS CoV-2
- PMID: 33629236
- PMCID: PMC7904240
- DOI: 10.1007/s10930-021-09967-8
A Biochemical Perspective of the Nonstructural Proteins (NSPs) and the Spike Protein of SARS CoV-2
Abstract
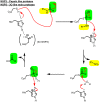
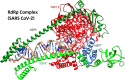
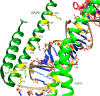
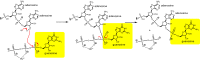
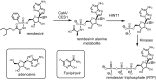
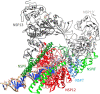
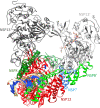
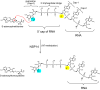
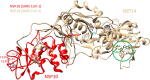
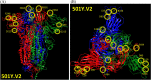
The global pandemic that shut down the world in 2020 was caused by the virus, SARS CoV-2. The chemistry of the various nonstructural proteins (NSP3, NSP5, NSP12, NSP13, NSP14, NSP15, NSP16) of SARS CoV-2 is discussed. Secondly, a recent major focus of this pandemic is the variant strains of SARS CoV-2 that are increasingly occurring and more transmissible. One strain, called "D614G", possesses a glycine (G) instead of an aspartate (D) at position 614 of the spike protein. Additionally, other emerging strains called "501Y.V1" and "501Y.V2" have several differences in the receptor binding domain of the spike protein (N501Y) as well as other locations. These structural changes may enhance the interaction between the spike protein and the ACE2 receptor of the host, increasing infectivity. The global pandemic caused by SARS CoV-2 is a rapidly evolving situation, emphasizing the importance of continuing the efforts to interrogate and understand this virus.
Keywords: Enzymes; Proteases; SARS CoV-2; Viral proteins.
Figures
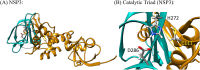
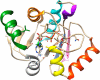

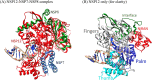














Similar articles
-
The Spike D614G mutation increases SARS-CoV-2 infection of multiple human cell types.Elife. 2021 Feb 11;10:e65365. doi: 10.7554/eLife.65365. Elife. 2021. PMID: 33570490 Free PMC article.
-
Characterization of SARS-CoV-2 Variants N501Y.V1 and N501Y.V2 Spike on Viral Infectivity.Front Cell Infect Microbiol. 2021 Oct 13;11:720357. doi: 10.3389/fcimb.2021.720357. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34722330 Free PMC article.
-
Transformations, Lineage Comparisons, and Analysis of Down-to-Up Protomer States of Variants of the SARS-CoV-2 Prefusion Spike Protein, Including the UK Variant B.1.1.7.Microbiol Spectr. 2021 Sep 3;9(1):e0003021. doi: 10.1128/Spectrum.00030-21. Epub 2021 Aug 4. Microbiol Spectr. 2021. PMID: 34346753 Free PMC article.
-
Angiotensin-Converting Enzyme 2 (ACE2) in the Pathogenesis of ARDS in COVID-19.Front Immunol. 2021 Dec 22;12:732690. doi: 10.3389/fimmu.2021.732690. eCollection 2021. Front Immunol. 2021. PMID: 35003058 Free PMC article. Review.
-
Structural basis of severe acute respiratory syndrome coronavirus 2 infection.Curr Opin HIV AIDS. 2021 Jan;16(1):74-81. doi: 10.1097/COH.0000000000000658. Curr Opin HIV AIDS. 2021. PMID: 33186231 Review.
Cited by
-
Beta, Delta, and Omicron, Deadliest Among SARS-CoV-2 Variants: A Computational Repurposing Approach.Evol Bioinform Online. 2023 Jul 11;19:11769343231182258. doi: 10.1177/11769343231182258. eCollection 2023. Evol Bioinform Online. 2023. PMID: 37457042 Free PMC article.
-
De novo ssRNA Aptamers against the SARS-CoV-2 Main Protease: In Silico Design and Molecular Dynamics Simulation.Int J Mol Sci. 2021 Jun 26;22(13):6874. doi: 10.3390/ijms22136874. Int J Mol Sci. 2021. PMID: 34206794 Free PMC article.
-
SARS-CoV-2 Non-Structural Proteins and Their Roles in Host Immune Evasion.Viruses. 2022 Sep 8;14(9):1991. doi: 10.3390/v14091991. Viruses. 2022. PMID: 36146796 Free PMC article. Review.
-
What do we know about the function of SARS-CoV-2 proteins?Front Immunol. 2023 Sep 18;14:1249607. doi: 10.3389/fimmu.2023.1249607. eCollection 2023. Front Immunol. 2023. PMID: 37790934 Free PMC article. Review.
-
Novel receptor, mutation, vaccine, and establishment of coping mode for SARS-CoV-2: current status and future.Front Microbiol. 2023 Aug 14;14:1232453. doi: 10.3389/fmicb.2023.1232453. eCollection 2023. Front Microbiol. 2023. PMID: 37645223 Free PMC article. Review.
References
-
- Mulligan MJ, et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;585:589–593. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

