Microfluidic Magneto Immunosensor for Rapid, High Sensitivity Measurements of SARS-CoV-2 Nucleocapsid Protein in Serum
- PMID: 33629833
- PMCID: PMC7931624
- DOI: 10.1021/acssensors.0c02561
Microfluidic Magneto Immunosensor for Rapid, High Sensitivity Measurements of SARS-CoV-2 Nucleocapsid Protein in Serum
Abstract
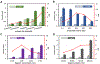
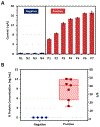
The COVID-19 pandemic has highlighted the importance and urgent need for rapid and accurate diagnostic tests for COVID-19 detection and screening. The objective of this work was to develop a simple immunosensor for rapid and high sensitivity measurements of SARS-CoV-2 nucleocapsid protein in serum. This assay is based on a unique sensing scheme utilizing dually-labeled magnetic nanobeads for immunomagnetic enrichment and signal amplification. This immunosensor is integrated onto a microfluidic chip, which offers the advantages of minimal sample and reagent consumption, simplified sample handling, and enhanced detection sensitivity. The functionality of this immunosensor was validated by using it to detect SARS-CoV-2 nucleocapsid protein, which could be detected at concentrations as low as 50 pg/mL in whole serum and 10 pg/mL in 5× diluted serum. We also adapted this assay onto a handheld smartphone-based diagnostic device that could detect SARS-CoV-2 nucleocapsid protein at concentrations as low as 230 pg/mL in whole serum and 100 pg/mL in 5× diluted serum. Lastly, we assessed the capability of this immunosensor to diagnose COVID-19 infection by testing clinical serum specimens, which revealed its ability to accurately distinguish PCR-positive COVID-19 patients from healthy, uninfected individuals based on SARS-CoV-2 nucleocapsid protein serum levels. To the best of our knowledge, this work is the first demonstration of rapid (<1 h) SARS-CoV-2 antigen quantification in whole serum samples. The ability to rapidly detect SARS-CoV-2 protein biomarkers with high sensitivity in very small (<50 μL) serum samples makes this platform a promising tool for point-of-care COVID-19 testing.
Keywords: COVID-19; SARS-CoV-2; electrochemical; immunosensor; microfluidic; nucleocapsid protein.
Figures



References
-
- COVID-19 Map - Johns Hopkins Coronavirus Resource Center https://coronavirus.jhu.edu/map.html (accessed Nov 8, 2020).
-
- Zhang W; Du RH; Li B; Zheng XS; Yang X. Lou; Hu B; Wang YY; Xiao GF; Yan B; Shi ZL; Zhou P. Molecular and Serological Investigation of 2019-NCoV Infected Patients: Implication of Multiple Shedding Routes. Emerg. Microbes Infect 2020, 9 (1), 386–389. 10.1080/22221751.2020.1729071. - DOI - PMC - PubMed
-
- Wölfel R; Corman VM; Guggemos W; Seilmaier M; Zange S; Müller MA; Niemeyer D; Jones TC; Vollmar P; Rothe C; Hoelscher M; Bleicker T; Brünink S; Schneider J; Ehmann R; Zwirglmaier K; Drosten C; Wendtner C Virological Assessment of Hospitalized Patients with COVID-2019. Nature 2020, 581 (7809), 465–469. 10.1038/s41586-020-2196-x. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

