Chronic HIV infection induces transcriptional and functional reprogramming of innate immune cells
- PMID: 33630761
- PMCID: PMC8119206
- DOI: 10.1172/jci.insight.145928
Chronic HIV infection induces transcriptional and functional reprogramming of innate immune cells
Abstract
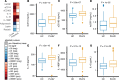
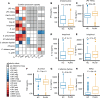
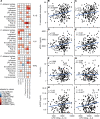
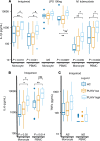
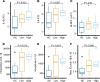
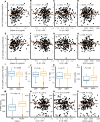
Chronic inflammation and immune dysfunction play a key role in the development of non-AIDS-related comorbidities. The aim of our study was to characterize the functional phenotype of immune cells in people living with HIV (PLHIV). We enrolled a cross-sectional cohort study of PLHIV on stable antiretroviral therapy and healthy controls. We assessed ex vivo cytokine production capacity and transcriptomics of monocytes and T cells upon bacterial, fungal, and viral stimulation. PLHIV exhibited an exacerbated proinflammatory profile in monocyte-derived cytokines, but not in lymphocyte-derived cytokines. Particularly, the production of the IL-1β to imiquimod, E. coli LPS, and Mycobacterium tuberculosis was increased, and this production correlated with plasma concentrations of high-sensitivity C-reactive protein and soluble CD14. This increase in monocyte responsiveness remained stable over time in subsequent blood sampling after more than 1 year. Transcriptome analyses confirmed priming of the monocyte IL-1β pathway, consistent with a monocyte-trained immunity phenotype. Increased plasma concentrations of β-glucan, a well-known inducer of trained immunity, were associated with increased innate cytokine responses. Monocytes of PLHIV exhibited a sustained proinflammatory immune phenotype with priming of the IL-1β pathway. Training of the innate immune system in PLHIV likely plays a role in long-term HIV complications and provides a promising therapeutic target for inflammation-related comorbidities.
Keywords: AIDS/HIV; Immunology; Innate immunity; Monocytes; Transcription.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

