Using a multiple-delivery-mode training approach to develop local capacity and infrastructure for advanced bioinformatics in Africa
- PMID: 33630830
- PMCID: PMC7906323
- DOI: 10.1371/journal.pcbi.1008640
Using a multiple-delivery-mode training approach to develop local capacity and infrastructure for advanced bioinformatics in Africa
Abstract
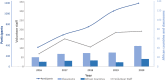
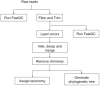
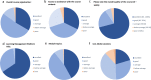
With more microbiome studies being conducted by African-based research groups, there is an increasing demand for knowledge and skills in the design and analysis of microbiome studies and data. However, high-quality bioinformatics courses are often impeded by differences in computational environments, complicated software stacks, numerous dependencies, and versions of bioinformatics tools along with a lack of local computational infrastructure and expertise. To address this, H3ABioNet developed a 16S rRNA Microbiome Intermediate Bioinformatics Training course, extending its remote classroom model. The course was developed alongside experienced microbiome researchers, bioinformaticians, and systems administrators, who identified key topics to address. Development of containerised workflows has previously been undertaken by H3ABioNet, and Singularity containers were used here to enable the deployment of a standard replicable software stack across different hosting sites. The pilot ran successfully in 2019 across 23 sites registered in 11 African countries, with more than 200 participants formally enrolled and 106 volunteer staff for onsite support. The pulling, running, and testing of the containers, software, and analyses on various clusters were performed prior to the start of the course by hosting classrooms. The containers allowed the replication of analyses and results across all participating classrooms running a cluster and remained available posttraining ensuring analyses could be repeated on real data. Participants thus received the opportunity to analyse their own data, while local staff were trained and supported by experienced experts, increasing local capacity for ongoing research support. This provides a model for delivering topic-specific bioinformatics courses across Africa and other remote/low-resourced regions which overcomes barriers such as inadequate infrastructures, geographical distance, and access to expertise and educational materials.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

