PFP-WGAN: Protein function prediction by discovering Gene Ontology term correlations with generative adversarial networks
- PMID: 33630862
- PMCID: PMC7906332
- DOI: 10.1371/journal.pone.0244430
PFP-WGAN: Protein function prediction by discovering Gene Ontology term correlations with generative adversarial networks
Abstract
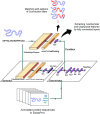
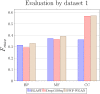
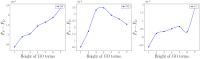
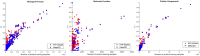
Understanding the functionality of proteins has emerged as a critical problem in recent years due to significant roles of these macro-molecules in biological mechanisms. However, in-laboratory techniques for protein function prediction are not as efficient as methods developed and processed for protein sequencing. While more than 70 million protein sequences are available today, only the functionality of around one percent of them are known. These facts have encouraged researchers to develop computational methods to infer protein functionalities from their sequences. Gene Ontology is the most well-known database for protein functions which has a hierarchical structure, where deeper terms are more determinative and specific. However, the lack of experimentally approved annotations for these specific terms limits the performance of computational methods applied on them. In this work, we propose a method to improve protein function prediction using their sequences by deeply extracting relationships between Gene Ontology terms. To this end, we construct a conditional generative adversarial network which helps to effectively discover and incorporate term correlations in the annotation process. In addition to the baseline algorithms, we compare our method with two recently proposed deep techniques that attempt to utilize Gene Ontology term correlations. Our results confirm the superiority of the proposed method compared to the previous works. Moreover, we demonstrate how our model can effectively help to assign more specific terms to sequences.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

