son is necessary for proper vertebrate blood development
- PMID: 33630943
- PMCID: PMC7906411
- DOI: 10.1371/journal.pone.0247489
son is necessary for proper vertebrate blood development
Abstract
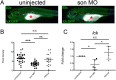
The gene SON is on human chromosome 21 (21q22.11) and is thought to be associated with hematopoietic disorders that accompany Down syndrome. Additionally, SON is an RNA splicing factor that plays a role in the transcription of leukemia-associated genes. Previously, we showed that mutations in SON cause malformations in human and zebrafish spines and brains during early embryonic development. To examine the role of SON in normal hematopoiesis, we reduced expression of the zebrafish homolog of SON in zebrafish at the single-cell developmental stage with specific morpholinos. In addition to the brain and spinal malformations we also observed abnormal blood cell levels upon son knockdown. We then investigated how blood production was altered when levels of son were reduced. Decreased levels of son resulted in lower amounts of red blood cells when visualized with lcr:GFP transgenic fish. There were also reduced thrombocytes seen with cd41:GFP fish, and myeloid cells when mpx:GFP fish were examined. We also observed a significant decrease in the quantity of T cells, visualized with lck:GFP fish. However, when we examined their hematopoietic stem and progenitor cells (HSPCs), we saw no difference in colony-forming capability. These studies indicate that son is essential for the proper differentiation of the innate and adaptive immune system, and further investigation determining the molecular pathways involved during blood development should elucidate important information about vertebrate HSPC generation, proliferation, and differentiation.
Conflict of interest statement
D.L.S. is a scientific consultant and has received compensation from Finless Foods, Inc. and Xytogen Biotech, Inc. This does not alter our adherence to PLOS ONE policies on sharing data and materials. All other authors declare no competing interests.
Figures




References
-
- Shivdasani RA, Orkin SH. The transcriptional control of hematopoiesis. Blood. 1996;87(10):4025–39. . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

