Gene expression underlying floral epidermal specialization in Aristolochia fimbriata (Aristolochiaceae)
- PMID: 33630993
- PMCID: PMC8103811
- DOI: 10.1093/aob/mcab033
Gene expression underlying floral epidermal specialization in Aristolochia fimbriata (Aristolochiaceae)
Abstract
Background and aims: The epidermis constitutes the outermost tissue of the plant body. Although it plays major structural, physiological and ecological roles in embryophytes, the molecular mechanisms controlling epidermal cell fate, differentiation and trichome development have been scarcely studied across angiosperms, and remain almost unexplored in floral organs.
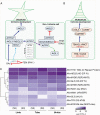
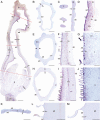
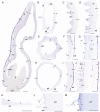
Methods: In this study, we assess the spatio-temporal expression patterns of GL2, GL3, TTG1, TRY, MYB5, MYB6, HDG2, MYB106-like, WIN1 and RAV1-like homologues in the magnoliid Aristolochia fimbriata (Aristolochiaceae) by using comparative RNA-sequencing and in situ hybridization assays.
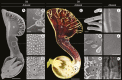
Key results: Genes involved in Aristolochia fimbriata trichome development vary depending on the organ where they are formed. Stem, leaf and pedicel trichomes recruit most of the transcription factors (TFs) described above. Conversely, floral trichomes only use a small subset of genes including AfimGL2, AfimRAV1-like, AfimWIN1, AfimMYB106-like and AfimHDG2. The remaining TFs, AfimTTG1, AfimGL3, AfimTRY, AfimMYB5 and AfimMYB6, are restricted to the abaxial (outer) and the adaxial (inner) pavement epidermal cells.
Conclusions: We re-evaluate the core genetic network shaping trichome fate in flowers of an early-divergent angiosperm lineage and show a morphologically diverse output with a simpler genetic mechanism in place when compared to the models Arabidopsis thaliana and Cucumis sativus. In turn, our results strongly suggest that the canonical trichome gene expression appears to be more conserved in vegetative than in floral tissues across angiosperms.
Keywords: Aristolochia; Piperales; epidermis development; multicellular trichomes; perianth; petaloid sepals.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Annals of Botany Company. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures




Similar articles
-
Comparative morphoanatomy and transcriptomic analyses reveal key factors controlling floral trichome development in Aristolochia (Aristolochiaceae).J Exp Bot. 2023 Nov 21;74(21):6588-6607. doi: 10.1093/jxb/erad345. J Exp Bot. 2023. PMID: 37656729
-
Flower Development and Perianth Identity Candidate Genes in the Basal Angiosperm Aristolochia fimbriata (Piperales: Aristolochiaceae).Front Plant Sci. 2015 Dec 11;6:1095. doi: 10.3389/fpls.2015.01095. eCollection 2015. Front Plant Sci. 2015. PMID: 26697047 Free PMC article.
-
APETALA3 and PISTILLATA homologs exhibit novel expression patterns in the unique perianth of Aristolochia (Aristolochiaceae).Evol Dev. 2004 Nov-Dec;6(6):449-58. doi: 10.1111/j.1525-142X.2004.04053.x. Evol Dev. 2004. PMID: 15509227
-
Flowering and trichome development share hormonal and transcription factor regulation.J Exp Bot. 2016 Mar;67(5):1209-19. doi: 10.1093/jxb/erv534. Epub 2015 Dec 17. J Exp Bot. 2016. PMID: 26685187 Review.
-
Trichome-Related Mutants Provide a New Perspective on Multicellular Trichome Initiation and Development in Cucumber (Cucumis sativus L).Front Plant Sci. 2016 Aug 10;7:1187. doi: 10.3389/fpls.2016.01187. eCollection 2016. Front Plant Sci. 2016. PMID: 27559338 Free PMC article. Review.
Cited by
-
De novo Assembly and Comparative Analyses of Mitochondrial Genomes in Piperales.Genome Biol Evol. 2023 Mar 3;15(3):evad041. doi: 10.1093/gbe/evad041. Genome Biol Evol. 2023. PMID: 36896589 Free PMC article.
-
Expression and Functional Studies of Leaf, Floral, and Fruit Developmental Genes in Non-model Species.Methods Mol Biol. 2023;2686:365-401. doi: 10.1007/978-1-0716-3299-4_19. Methods Mol Biol. 2023. PMID: 37540370
-
Uncovering developmental diversity in the field.Development. 2024 Oct 15;151(20):dev203084. doi: 10.1242/dev.203084. Epub 2024 Aug 19. Development. 2024. PMID: 39158021 Free PMC article.
References
-
- Aliscioni SS, Achler AP, Torretta JP. 2017. Floral anatomy, micromorphology and visitor insects in three species of Aristolochia L. (Aristolochiaceae). New Zealand Journal of Botany 55: 496–513.
-
- Ambrose BA, Lerner DR, Ciceri P, Padilla CM, Yanofsky MF, Schmidt RJ. 2000. Molecular and genetic analyses of the Silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Molecular Cell 5: 569–579. - PubMed
-
- Balkunde R, Pesch M, Hülskamp M. 2011. Trichome patterning in Arabidopsis thaliana: from genetic to molecular models. Current Topics in Developmental Biology 91: 299–321. - PubMed
-
- Baumann K, Perez-Rodriguez M, Bradley D, et al. . 2007. Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development (Cambridge, England) 134: 1691–1701. - PubMed
-
- Bernhardt C, Lee MM, Gonzalez A, Zhang F, Lloyd A, Schiefelbein J. 2003. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development (Cambridge, England) 130: 6431–6439. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

