Revealing Tissue-Specific SARS-CoV-2 Infection and Host Responses using Human Stem Cell-Derived Lung and Cerebral Organoids
- PMID: 33631122
- PMCID: PMC7879814
- DOI: 10.1016/j.stemcr.2021.02.005
Revealing Tissue-Specific SARS-CoV-2 Infection and Host Responses using Human Stem Cell-Derived Lung and Cerebral Organoids
Abstract
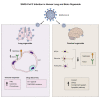
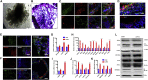
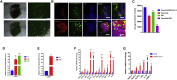
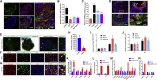
COVID-19 is a transmissible respiratory disease caused by a novel coronavirus, SARS-CoV-2, and has become a global health emergency. There is an urgent need for robust and practical in vitro model systems to investigate viral pathogenesis. Here, we generated human induced pluripotent stem cell (iPSC)-derived lung organoids (LORGs), cerebral organoids (CORGs), neural progenitor cells (NPCs), neurons, and astrocytes. LORGs containing epithelial cells, alveolar types 1 and 2, highly express ACE2 and TMPRSS2 and are permissive to SARS-CoV-2 infection. SARS-CoV-2 infection induces interferons, cytokines, and chemokines and activates critical inflammasome pathway genes. Spike protein inhibitor, EK1 peptide, and TMPRSS2 inhibitors (camostat/nafamostat) block viral entry in LORGs. Conversely, CORGs, NPCs, astrocytes, and neurons express low levels of ACE2 and TMPRSS2 and correspondingly are not highly permissive to SARS-CoV-2 infection. Infection in neuronal cells activates TLR3/7, OAS2, complement system, and apoptotic genes. These findings will aid in understanding COVID-19 pathogenesis and facilitate drug discovery.
Keywords: ACE2; SARS-CoV-2; TMPRSS2; cerebral organoids; host-pathogen interactions; iPSCs; lungs organoids.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
T.M.R. is a founder of ViRx Pharmaceuticals and has an equity interest in the company. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

