Pervasive transmission of E484K and emergence of VUI-NP13L with evidence of SARS-CoV-2 co-infection events by two different lineages in Rio Grande do Sul, Brazil
- PMID: 33631222
- PMCID: PMC7898980
- DOI: 10.1016/j.virusres.2021.198345
Pervasive transmission of E484K and emergence of VUI-NP13L with evidence of SARS-CoV-2 co-infection events by two different lineages in Rio Grande do Sul, Brazil
Abstract
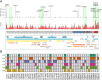
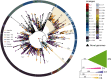
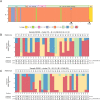
Emergence of novel SARS-CoV-2 lineages are under the spotlight of the media, scientific community and governments. Recent reports of novel variants in the United Kingdom, South Africa and Brazil (B.1.1.28-E484K) have raised intense interest because of a possible higher transmission rate or resistance to the novel vaccines. Nevertheless, the spread of B.1.1.28 (E484K) and other variants in Brazil is still unknown. In this work, we investigated the population structure and genomic complexity of SARS-CoV-2 in Rio Grande do Sul, the southernmost state in Brazil. Most samples sequenced belonged to the B.1.1.28 (E484K) lineage, demonstrating its widespread dispersion. We were the first to identify two independent events of co-infection caused by the occurrence of B.1.1.28 (E484K) with either B.1.1.248 or B.1.91 lineages. Also, clustering analysis revealed the occurrence of a novel cluster of samples circulating in the state (named VUI-NP13L) characterized by 12 lineage-defining mutations. In light of the evidence for E484K dispersion, co-infection and emergence of VUI-NP13 L in Rio Grande do Sul, we reaffirm the importance of establishing strict and effective social distancing measures to counter the spread of potentially more hazardous SARS-CoV-2 strains.
Keywords: COVID-19; Co-infection; E484K; Genomic surveillance; Mutation; P13L.
Copyright © 2021. Published by Elsevier B.V.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
References
-
- Banu S., Jolly B., Mukherjee P., Singh P., Khan S., Zaveri L., Shambhavi S., Gaur N., Reddy S., Kaveri K., Srinivasan S., Gopal D.R., Siva A.B., Thangaraj K., Tallapaka K.B., Mishra R.K., Scaria V., Sowpati D.T. A distinct phylogenetic cluster of Indian severe acute respiratory syndrome coronavirus 2 isolates. Open Forum Infect. Dis. 2020;7 ofaa434. - PMC - PubMed
-
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25 - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

