Highly Multiplexed Immunohistochemical MALDI-MS Imaging of Biomarkers in Tissues
- PMID: 33631930
- PMCID: PMC8033562
- DOI: 10.1021/jasms.0c00473
Highly Multiplexed Immunohistochemical MALDI-MS Imaging of Biomarkers in Tissues
Abstract
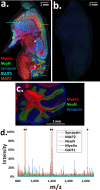
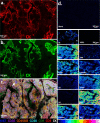
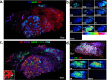
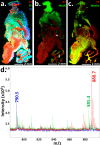
Immunohistochemistry (IHC) combined with fluorescence microscopy provides an important and widely used tool for researchers and pathologists to image multiple biomarkers in tissue specimens. However, multiplex IHC using standard fluorescence microscopy is generally limited to 3-5 different biomarkers, with hyperspectral or multispectral methods limited to 8. We report the development of a new technology based on novel photocleavable mass-tags (PC-MTs) for facile antibody labeling, which enables highly multiplexed IHC based on MALDI mass spectrometric imaging (MALDI-IHC). This approach significantly exceeds the multiplexity of both fluorescence- and previous cleavable mass-tag-based methods. Up to 12-plex MALDI-IHC was demonstrated on mouse brain, human tonsil, and breast cancer tissues specimens, reflecting the known molecular composition, anatomy, and pathology of the targeted biomarkers. Novel dual-labeled fluorescent PC-MT antibodies and label-free small-molecule mass spectrometric imaging greatly extend the capability of this new approach. MALDI-IHC shows promise for use in the fields of tissue pathology, tissue diagnostics, therapeutics, and precision medicine.
Keywords: bead-arrays; immunofluorescence; immunohistochemistry; in situ hybridization; mass spectrometric imaging; mass spectrometry; mass-tags; matrix-assisted laser desorption/ionization; multiplexing; pathology; photocleavable; tissue diagnostics.
Conflict of interest statement
The authors declare the following competing financial interest(s): G.Y., Z.L., K.J.R., and M.J.L. are current employees of AmberGen, Inc., 313 Pleasant Street, Watertown, MA 02472, USA. AmberGen, Inc. has filed patent applications on aspects of this work.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

