Molecular profile of KRAS G12C-mutant colorectal and non-small-cell lung cancer
- PMID: 33632153
- PMCID: PMC7905642
- DOI: 10.1186/s12885-021-07884-8
Molecular profile of KRAS G12C-mutant colorectal and non-small-cell lung cancer
Abstract
Background: KRAS is the most frequently mutated oncogene in cancer, however efforts to develop targeted therapies have been largely unsuccessful. Recently, two small-molecule inhibitors, AMG 510 and MRTX849, have shown promising activity in KRAS G12C-mutant solid tumors. The current study aims to assess the molecular profile of KRAS G12C in colorectal (CRC) and non-small-cell lung cancer (NSCLC) tested in a clinical certified laboratory.
Methods: CRC and NSCLC samples submitted for KRAS testing between 2017 and 2019 were reviewed. CRC samples were tested for KRAS and NRAS by pyrosequencing, while NSCLC samples were submitted to next generation sequencing of KRAS, NRAS, EGFR, and BRAF.
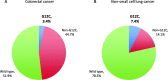
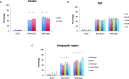
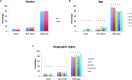
Results: The dataset comprised 4897 CRC and 4686 NSCLC samples. Among CRC samples, KRAS was mutated in 2354 (48.1%). Most frequent codon 12 mutations were G12D in 731 samples (14.9%) and G12V in 522 (10.7%), followed by G12C in 167 (3.4%). KRAS mutations were more frequent in females than males (p = 0.003), however this difference was exclusive of non-G12C mutants (p < 0.001). KRAS mutation frequency was lower in the South and North regions (p = 0.003), but again KRAS G12C did not differ significantly (p = 0.80). In NSCLC, KRAS mutations were found in 1004 samples (21.4%). As opposed to CRC samples, G12C was the most common mutation in KRAS, in 346 cases (7.4%). The frequency of KRAS G12C was higher in the South and Southeast regions (p = 0.012), and lower in patients younger than 50 years (p < 0.001). KRAS G12C mutations were largely mutually exclusive with other driver mutations; only 11 NSCLC (3.2%) and 1 CRC (0.6%) cases had relevant co-mutations.
Conclusions: KRAS G12C presents in frequencies higher than several other driver mutations, and may represent a large volume of patients in absolute numbers. KRAS testing should be considered in all CRC and NSCLC patients, independently of clinical or demographic characteristics.
Keywords: Colorectal neoplasms; KRAS; Lung neoplasms; Molecular diagnostics; Molecular targeted therapy.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Heinrich MC, Corless CL, Demetri GD, Blanke CD, von Mehren M, Joensuu H, McGreevey LS, Chen CJ, Van den Abbeele AD, Druker BJ, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21(23):4342–4349. doi: 10.1200/JCO.2003.04.190. - DOI - PubMed
-
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

