Melatonin promotes bone marrow mesenchymal stem cell osteogenic differentiation and prevents osteoporosis development through modulating circ_0003865 that sponges miR-3653-3p
- PMID: 33632317
- PMCID: PMC7908669
- DOI: 10.1186/s13287-021-02224-w
Melatonin promotes bone marrow mesenchymal stem cell osteogenic differentiation and prevents osteoporosis development through modulating circ_0003865 that sponges miR-3653-3p
Abstract
Background: Little is known about the implications of circRNAs in the effects of melatonin (MEL) on bone marrow mesenchymal stem cell (BMSC) osteogenic differentiation and osteoporosis (OP) progression. The aim of our study was to investigate circRNAs in MEL-regulated BMSC differentiation and OP progression.
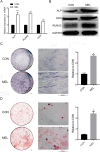
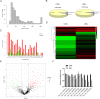
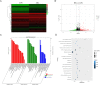
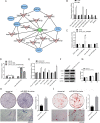
Methods: BMSC osteogenic differentiation was measured by qRT-PCR, western blot (WB), Alizarin Red, and alkaline phosphatase (ALP) staining. Differential circRNA and mRNA profiles of BMSCs treated by MEL were characterized by deep sequencing, followed by validation using RT-PCR, Sanger sequencing, and qRT-PCR. Silencing and overexpression of circ_0003865 were conducted for functional investigations. The sponged microRNAs and targeted mRNAs were predicted by bioinformatics and validated by qRT-PCR, RNA pull-down, and dual-luciferase reporter assay. The function of miR-3653-3p and circ_0003865/miR-3653-3p/growth arrest-specific gene 1 (GAS1) cascade was validated for the osteogenic differentiation of BMSCs by CCK-8, qRT-PCR, WB, Alizarin Red, and ALP staining. The effects of circ_0003865 on OP development were tested in murine OP model.
Results: MEL promoted osteogenic differentiation of BMSCs. RNA sequencing revealed significant alterations in circRNA and mRNA profiles associated with multiple biological processes and signaling pathways. Circ_0003865 expression in BMSCs was significantly decreased by MEL treatment. Silencing of circ_0003865 had no effect on proliferation while promoted osteogenic differentiation of BMSCs. Overexpression of circ_0003865 abrogated the promotion of BMSC osteogenic differentiation induced by MEL, but proliferation of BMSCs induced by MEL had no change whether circ_0003865 was overexpression or not. Furthermore, circ_0003865 sponged miR-3653-3p to promote GAS1 expression in BMSCs. BMSC osteogenic differentiation was enhanced by miR-3653-3p overexpression while BMSC proliferation was not affected. By contrast, miR-3653-3p silencing mitigated the promoted BMSC osteogenic differentiation caused by circ_0003865 silencing, but had no effect on proliferation. Finally, circ_0003865 silencing repressed OP development in mouse model.
Conclusion: MEL promotes BMSC osteogenic differentiation and inhibits OP pathogenesis by suppressing the expression of circ_0003865, which regulates GAS1 gene expression via sponging miR-3653-3p.
Keywords: BMSCs; Melatonin; Osteogenic differentiation; Osteoporosis; circ_0003865; miR-3653-3p.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
A novel link between melatonin and circ_0005753/PTBP1/TXNIP regulatory network in the modulation of osteogenic potential in mesenchymal stem cells.Chem Biol Drug Des. 2024 Jan;103(1):e14380. doi: 10.1111/cbdd.14380. Epub 2023 Oct 27. Chem Biol Drug Des. 2024. PMID: 37890873
-
circ-Iqsec1 induces bone marrow-derived mesenchymal stem cell (BMSC) osteogenic differentiation through the miR-187-3p/Satb2 signaling pathway.Arthritis Res Ther. 2022 Dec 14;24(1):273. doi: 10.1186/s13075-022-02964-x. Arthritis Res Ther. 2022. PMID: 36517907 Free PMC article.
-
Circ_0019693 promotes osteogenic differentiation of bone marrow mesenchymal stem cell and enhances osteogenesis-coupled angiogenesis via regulating microRNA-942-5p-targeted purkinje cell protein 4 in the development of osteoporosis.Bioengineered. 2022 Feb;13(2):2181-2193. doi: 10.1080/21655979.2021.2023982. Bioengineered. 2022. PMID: 35030971 Free PMC article.
-
Research progress on the role of lncRNA-miRNA networks in regulating adipogenic and osteogenic differentiation of bone marrow mesenchymal stem cells in osteoporosis.Front Endocrinol (Lausanne). 2023 Aug 14;14:1210627. doi: 10.3389/fendo.2023.1210627. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37645421 Free PMC article. Review.
-
Plasma proteins, circulating metabolites mediate causal inference studies on the effect of gut bacteria on the risk of osteoporosis development.Ageing Res Rev. 2024 Nov;101:102479. doi: 10.1016/j.arr.2024.102479. Epub 2024 Aug 28. Ageing Res Rev. 2024. PMID: 39214170 Review.
Cited by
-
Melatonin Inhibits Osteoclastogenesis and Osteolytic Bone Metastasis: Implications for Osteoporosis.Int J Mol Sci. 2021 Aug 30;22(17):9435. doi: 10.3390/ijms22179435. Int J Mol Sci. 2021. PMID: 34502344 Free PMC article. Review.
-
Aberrant Tryptophan Metabolism Manipulates Osteochondral Homeostasis.Research (Wash D C). 2025 Jun 10;8:0728. doi: 10.34133/research.0728. eCollection 2025. Research (Wash D C). 2025. PMID: 40496774 Free PMC article. Review.
-
The roles of circRNA-miRNA-mRNA networks in the development and treatment of osteoporosis.Front Endocrinol (Lausanne). 2022 Aug 5;13:945310. doi: 10.3389/fendo.2022.945310. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35992137 Free PMC article. Review.
-
Aspirin attenuates the detrimental effects of TNF-α on BMMSC stemness by modulating the YAP-SMAD7 axis.Mol Med. 2024 Aug 16;30(1):126. doi: 10.1186/s10020-024-00890-z. Mol Med. 2024. PMID: 39152406 Free PMC article.
-
Aspirin reverses inflammatory suppression of chondrogenesis by stabilizing YAP.Cell Prolif. 2023 Apr;56(4):e13380. doi: 10.1111/cpr.13380. Epub 2022 Dec 10. Cell Prolif. 2023. PMID: 36495056 Free PMC article.
References
-
- Manolagas SC. Birth and death of bone cells: basic regulatory mechanisms and implications for the pathogenesis and treatment of osteoporosis. Endocr Rev. 2000;21(1):115–137. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

