Shulin packages axonemal outer dynein arms for ciliary targeting
- PMID: 33632841
- PMCID: PMC7116892
- DOI: 10.1126/science.abe0526
Shulin packages axonemal outer dynein arms for ciliary targeting
Abstract
The main force generators in eukaryotic cilia and flagella are axonemal outer dynein arms (ODAs). During ciliogenesis, these ~1.8-megadalton complexes are assembled in the cytoplasm and targeted to cilia by an unknown mechanism. Here, we used the ciliate Tetrahymena to identify two factors (Q22YU3 and Q22MS1) that bind ODAs in the cytoplasm and are required for ODA delivery to cilia. Q22YU3, which we named Shulin, locked the ODA motor domains into a closed conformation and inhibited motor activity. Cryo-electron microscopy revealed how Shulin stabilized this compact form of ODAs by binding to the dynein tails. Our findings provide a molecular explanation for how newly assembled dyneins are packaged for delivery to the cilia.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures
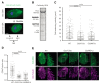
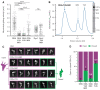

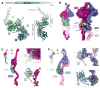
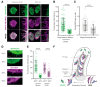
References
-
- Goodenough UW, Heuser JE. Outer and inner dynein arms of cilia and flagella. Cell. 1985;41:341–342. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

