Recursive ensemble feature selection provides a robust mRNA expression signature for myalgic encephalomyelitis/chronic fatigue syndrome
- PMID: 33633136
- PMCID: PMC7907358
- DOI: 10.1038/s41598-021-83660-9
Recursive ensemble feature selection provides a robust mRNA expression signature for myalgic encephalomyelitis/chronic fatigue syndrome
Abstract
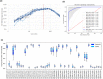
Myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) is a chronic disorder characterized by disabling fatigue. Several studies have sought to identify diagnostic biomarkers, with varying results. Here, we innovate this process by combining both mRNA expression and DNA methylation data. We performed recursive ensemble feature selection (REFS) on publicly available mRNA expression data in peripheral blood mononuclear cells (PBMCs) of 93 ME/CFS patients and 25 healthy controls, and found a signature of 23 genes capable of distinguishing cases and controls. REFS highly outperformed other methods, with an AUC of 0.92. We validated the results on a different platform (AUC of 0.95) and in DNA methylation data obtained from four public studies on ME/CFS (99 patients and 50 controls), identifying 48 gene-associated CpGs that predicted disease status as well (AUC of 0.97). Finally, ten of the 23 genes could be interpreted in the context of the derailed immune system of ME/CFS.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

