CIBERSORT analysis of TCGA and METABRIC identifies subgroups with better outcomes in triple negative breast cancer
- PMID: 33633150
- PMCID: PMC7907367
- DOI: 10.1038/s41598-021-83913-7
CIBERSORT analysis of TCGA and METABRIC identifies subgroups with better outcomes in triple negative breast cancer
Abstract
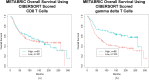
Studies have shown that the presence of tumor infiltrating lymphocytes (TILs) in Triple Negative Breast Cancer (TNBC) is associated with better prognosis. However, the molecular mechanisms underlying these immune cell differences are not well delineated. In this study, analysis of hematoxylin and eosin images from The Cancer Genome Atlas (TCGA) breast cancer cohort failed to show a prognostic benefit of TILs in TNBC, whereas CIBERSORT analysis, which quantifies the proportion of each immune cell type, demonstrated improved overall survival in TCGA TNBC samples with increased CD8 T cells or CD8 plus CD4 memory activated T cells and in Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) TNBC samples with increased gamma delta T cells. Twenty-five genes showed mutational frequency differences between the TCGA high and low T cell groups, and many play important roles in inflammation or immune evasion (ATG2B, HIST1H2BC, PKD1, PIKFYVE, TLR3, NOTCH3, GOLGB1, CREBBP). Identification of these mutations suggests novel mechanisms by which the cancer cells attract immune cells and by which they evade or dampen the immune system during the cancer immunoediting process. This study suggests that integration of mutations with CIBERSORT analysis could provide better prediction of outcomes and novel therapeutic targets in TNBC cases.
Conflict of interest statement
The authors declare no competing interests.
Figures


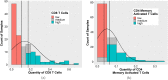
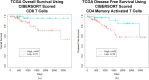
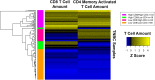


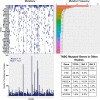

Similar articles
-
A Machine Learning Model to Predict the Triple Negative Breast Cancer Immune Subtype.Front Immunol. 2021 Sep 17;12:749459. doi: 10.3389/fimmu.2021.749459. eCollection 2021. Front Immunol. 2021. PMID: 34603338 Free PMC article.
-
Insulin-like growth factor 2 receptor is a key immune-related gene that is correlated with a poor prognosis in patients with triple-negative breast cancer: A bioinformatics analysis.Front Oncol. 2022 Oct 18;12:871786. doi: 10.3389/fonc.2022.871786. eCollection 2022. Front Oncol. 2022. PMID: 36330486 Free PMC article.
-
Tumor-infiltrating B cells and T cells correlate with postoperative prognosis in triple-negative carcinoma of the breast.BMC Cancer. 2021 Mar 16;21(1):286. doi: 10.1186/s12885-021-08009-x. BMC Cancer. 2021. PMID: 33726701 Free PMC article.
-
Prognostic value of tumor-infiltrating lymphocytes in patients with triple-negative breast cancer: a systematic review and meta-analysis.BMC Cancer. 2020 Mar 4;20(1):179. doi: 10.1186/s12885-020-6668-z. BMC Cancer. 2020. PMID: 32131780 Free PMC article.
-
Role of inflammatory infiltrates in triple negative breast cancer.J Clin Pathol. 2015 Jul;68(7):506-10. doi: 10.1136/jclinpath-2015-202944. Epub 2015 Mar 6. J Clin Pathol. 2015. PMID: 25750267 Review.
Cited by
-
Construction and Clinical Translation of Causal Pan-Cancer Gene Score Across Cancer Types.Front Genet. 2021 Dec 23;12:784775. doi: 10.3389/fgene.2021.784775. eCollection 2021. Front Genet. 2021. PMID: 35003220 Free PMC article.
-
Comprehensive bioinformatics analytics and in vivo validation reveal SLC31A1 as an emerging diagnostic biomarker for acute myocardial infarction.Aging (Albany NY). 2024 May 6;16(9):8361-8377. doi: 10.18632/aging.205199. Epub 2024 May 6. Aging (Albany NY). 2024. PMID: 38713173 Free PMC article.
-
Identifying biomarkers of endoplasmic reticulum stress and analyzing immune cell infiltration in chronic obstructive pulmonary disease using machine learning.Front Med (Lausanne). 2024 Nov 22;11:1462868. doi: 10.3389/fmed.2024.1462868. eCollection 2024. Front Med (Lausanne). 2024. PMID: 39650190 Free PMC article.
-
Exploring the Interconnections Between Mitochondrial Dysfunction and Polycystic Ovary Syndrome: A Comprehensive Integrated Analysis.Biochem Genet. 2025 Apr 21. doi: 10.1007/s10528-025-11104-4. Online ahead of print. Biochem Genet. 2025. PMID: 40259200
-
Triple negative breast cancer: Immunogenicity, tumor microenvironment, and immunotherapy.Front Genet. 2023 Jan 12;13:1095839. doi: 10.3389/fgene.2022.1095839. eCollection 2022. Front Genet. 2023. PMID: 36712858 Free PMC article. Review.
References
-
- Loi S, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02–98. J. Clin. Oncol. 2013;31:860–7. doi: 10.1200/JCO.2011.41.0902. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

