An NKX2-1GFP and TP63tdTomato dual fluorescent reporter for the investigation of human lung basal cell biology
- PMID: 33633173
- PMCID: PMC7907081
- DOI: 10.1038/s41598-021-83825-6
An NKX2-1GFP and TP63tdTomato dual fluorescent reporter for the investigation of human lung basal cell biology
Abstract
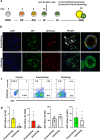
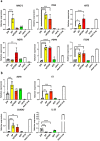
Basal cells are multipotent stem cells responsible for the repair and regeneration of all the epithelial cell types present in the proximal lung. In mice, the elusive origins of basal cells and their contribution to lung development were recently revealed by high-resolution, lineage tracing studies. It however remains unclear if human basal cells originate and participate in lung development in a similar fashion, particularly with mounting evidence for significant species-specific differences in this process. To address this outstanding question, in the last several years differentiation protocols incorporating human pluripotent stem cells (hPSC) have been developed to produce human basal cells in vitro with varying efficiencies. To facilitate this endeavour, we introduced tdTomato into the human TP63 gene, whose expression specifically labels basal cells, in the background of a previously described hPSC line harbouring an NKX2-1GFP reporter allele. The functionality and specificity of the NKX2-1GFP;TP63tdTomato hPSC line was validated by directed differentiation into lung progenitors as well as more specialised lung epithelial subtypes using an organoid platform. This dual fluorescent reporter hPSC line will be useful for tracking, isolating and expanding basal cells from heterogenous differentiation cultures for further study.
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Derivation of Airway Basal Stem Cells from Human Pluripotent Stem Cells.Cell Stem Cell. 2021 Jan 7;28(1):79-95.e8. doi: 10.1016/j.stem.2020.09.017. Epub 2020 Oct 23. Cell Stem Cell. 2021. PMID: 33098807 Free PMC article.
-
Influence of mesenchymal and biophysical components on distal lung organoid differentiation.Stem Cell Res Ther. 2024 Sep 2;15(1):273. doi: 10.1186/s13287-024-03890-2. Stem Cell Res Ther. 2024. PMID: 39218985 Free PMC article.
-
Pluripotent Stem Cell-Derived Cerebral Organoids Reveal Human Oligodendrogenesis with Dorsal and Ventral Origins.Stem Cell Reports. 2019 May 14;12(5):890-905. doi: 10.1016/j.stemcr.2019.04.011. Stem Cell Reports. 2019. PMID: 31091434 Free PMC article.
-
A critical look: Challenges in differentiating human pluripotent stem cells into desired cell types and organoids.Wiley Interdiscip Rev Dev Biol. 2020 May;9(3):e368. doi: 10.1002/wdev.368. Epub 2019 Nov 19. Wiley Interdiscip Rev Dev Biol. 2020. PMID: 31746148 Free PMC article. Review.
-
Tumor imaging with multicolor fluorescent protein expression.Int J Clin Oncol. 2011 Apr;16(2):84-91. doi: 10.1007/s10147-011-0201-y. Epub 2011 Feb 25. Int J Clin Oncol. 2011. PMID: 21347627 Review.
Cited by
-
Reconstructing the pulmonary niche with stem cells: a lung story.Stem Cell Res Ther. 2022 Apr 11;13(1):161. doi: 10.1186/s13287-022-02830-2. Stem Cell Res Ther. 2022. PMID: 35410254 Free PMC article. Review.
-
Distinctive field effects of smoking and lung cancer case-control status on bronchial basal cell growth and signaling.Respir Res. 2024 Aug 19;25(1):317. doi: 10.1186/s12931-024-02924-w. Respir Res. 2024. PMID: 39160511 Free PMC article.
References
-
- Society, E. R. Forum of International Respiratory Societies. The Global Impact of Respiratory Disease – Second Edition. Sheffield,. (2017).
-
- Bowden DH. Cell turnover in the lung. Am. Rev. Respir. Dis. 1983;128:2. - PubMed
-
- Kauffman SL. Cell proliferation in the mammalian lung. Int. Rev. Exp. Pathol. 1980;22:131–191. - PubMed
-
- Adamson IYR, Bowden DH. The type 2 cell as progenitor of alveolar epithelial regeneration: A cytodynamic study in mice after exposure to oxygen. Lab. Investig. 1974;30:35–42. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

