Exploring the conservation of Alzheimer-related pathways between H. sapiens and C. elegans: a network alignment approach
- PMID: 33633188
- PMCID: PMC7907373
- DOI: 10.1038/s41598-021-83892-9
Exploring the conservation of Alzheimer-related pathways between H. sapiens and C. elegans: a network alignment approach
Abstract
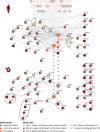
Alzheimer disease (AD) is a neurodegenerative disorder with an -as of yet- unclear etiology and pathogenesis. Research to unveil disease processes underlying AD often relies on the use of neurodegenerative disease model organisms, such as Caenorhabditis elegans. This study sought to identify biological pathways implicated in AD that are conserved in Homo sapiens and C. elegans. Protein-protein interaction networks were assembled for amyloid precursor protein (APP) and Tau in H. sapiens-two proteins whose aggregation is a hallmark in AD-and their orthologs APL-1 and PTL-1 for C. elegans. Global network alignment was used to compare these networks and determine similar, likely conserved, network regions. This comparison revealed that two prominent pathways, the APP-processing and the Tau-phosphorylation pathways, are highly conserved in both organisms. While the majority of interactions between proteins in those pathways are known to be associated with AD in human, they remain unexamined in C. elegans, signifying the need for their further investigation. In this work, we have highlighted conserved interactions related to AD in humans and have identified specific proteins that can act as targets for experimental studies in C. elegans, aiming to uncover the underlying mechanisms of AD.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

