Evolutionary conservation of the DRACH signatures of potential N6-methyladenosine (m6A) sites among influenza A viruses
- PMID: 33633224
- PMCID: PMC7907337
- DOI: 10.1038/s41598-021-84007-0
Evolutionary conservation of the DRACH signatures of potential N6-methyladenosine (m6A) sites among influenza A viruses
Abstract
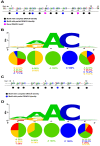
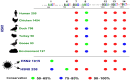
The addition of a methyl group to the N6-position of adenosine (m6A) is considered one of the most prevalent internal post-transcriptional modifications and is attributed to virus replication and cell biology. Viral epitranscriptome sequencing analysis has revealed that hemagglutinin (HA) mRNA of H1N1 carry eight m6A sites which are primarily enriched in 5'-DRACH-3' sequence motif. Herein, a large-scale comparative m6A analysis was conducted to investigate the conservation patterns of the DRACH motifs that corresponding to the reference m6A sites among influenza A viruses. A total of 70,030 complete HA sequences that comprise all known HA subtypes (H1-18) collected over several years, countries, and affected host species were analysed on both mRNA and vRNA strands. The bioinformatic analysis revealed the highest degree of DRACHs conservation among all H1 sequences that clustered largely in the middle and in the vicinity to 3' end with at least four DRACH motifs were conserved in all mRNA sequences. The major HA-containing subtypes displayed a modest DRACH motif conservation located either in the middle region of HA transcript (H3) or at the 3' end (H5) or were distributed across the length of HA sequence (H9). The lowest conservation was demonstrated in HA subtypes that infect mostly the wild type avian species and bats. Interestingly, the total number and the conserved DRACH motifs in the vRNA were found to be much lower than those observed in the mRNA. Collectively, the identification of putative m6A topology provides a foundation for the future intervention of influenza infection, replication, and pathobiology in susceptible hosts.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

