Comparative genomics of MRSA strains from human and canine origins reveals similar virulence gene repertoire
- PMID: 33633263
- PMCID: PMC7907190
- DOI: 10.1038/s41598-021-83993-5
Comparative genomics of MRSA strains from human and canine origins reveals similar virulence gene repertoire
Abstract
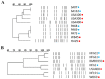
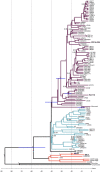
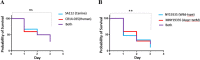
Methicillin-resistant Staphylococcus aureus (MRSA) is an important pathogen associated with a wide variety of infections in humans. The ability of MRSA to infect companion animals has gained increasing attention in the scientific literature. In this study, 334 dogs were screened for MRSA in two cities located in Rio de Janeiro State. The prevalence of MRSA in dogs was 2.7%. Genotyping revealed isolates from sequence types (ST) 1, 5, 30, and 239 either colonizing or infecting dogs. The genome of the canine ST5 MRSA (strain SA112) was compared with ST5 MRSA from humans-the main lineage found in Rio de Janeiro hospitals-to gain insights in the origin of this dog isolate. Phylogenetic analysis situated the canine genome and human strain CR14-035 in the same clade. Comparative genomics revealed similar virulence profiles for SA112 and CR14-035. Both genomes carry S. aureus genomic islands νSAα, νSAβ, and νSAγ. The virulence potential of the canine and human strains was similar in a Caenorhabditis elegans model. Together, these results suggest a potential of canine MRSA to infect humans and vice versa. The circulation in community settings of a MRSA lineage commonly found in hospitals is an additional challenge for public health surveillance authorities.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- van Balen J, et al. Presence, distribution, and molecular epidemiology of methicillin-resistant Staphylococcus aureus in a small animal teaching hospital: A year-long active surveillance targeting dogs and their environment. Vector Borne Zoonot. Dis. 2013;13:299–311. doi: 10.1089/vbz.2012.1142. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

