Role of the V1G1 subunit of V-ATPase in breast cancer cell migration
- PMID: 33633298
- PMCID: PMC7907067
- DOI: 10.1038/s41598-021-84222-9
Role of the V1G1 subunit of V-ATPase in breast cancer cell migration
Abstract
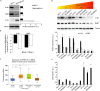
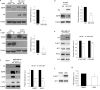
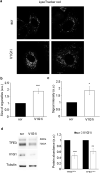
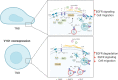
V-ATPase is a large multi-subunit complex that regulates acidity of intracellular compartments and of extracellular environment. V-ATPase consists of several subunits that drive specific regulatory mechanisms. The V1G1 subunit, a component of the peripheral stalk of the pump, controls localization and activation of the pump on late endosomes and lysosomes by interacting with RILP and RAB7. Deregulation of some subunits of the pump has been related to tumor invasion and metastasis formation in breast cancer. We observed a decrease of V1G1 and RAB7 in highly invasive breast cancer cells, suggesting a key role of these proteins in controlling cancer progression. Moreover, in MDA-MB-231 cells, modulation of V1G1 affected cell migration and matrix metalloproteinase activation in vitro, processes important for tumor formation and dissemination. In these cells, characterized by high expression of EGFR, we demonstrated that V1G1 modulates EGFR stability and the EGFR downstream signaling pathways that control several factors required for cell motility, among which RAC1 and cofilin. In addition, we showed a key role of V1G1 in the biogenesis of endosomes and lysosomes. Altogether, our data describe a new molecular mechanism, controlled by V1G1, required for cell motility and that promotes breast cancer tumorigenesis.
Conflict of interest statement
The authors declare no conflict of interest. The funder had no role in the design of the study, in the collection, analyses or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures



Similar articles
-
RILP regulates vacuolar ATPase through interaction with the V1G1 subunit.J Cell Sci. 2014 Jun 15;127(Pt 12):2697-708. doi: 10.1242/jcs.142604. Epub 2014 Apr 24. J Cell Sci. 2014. PMID: 24762812
-
A new V-ATPase regulatory mechanism mediated by the Rab interacting lysosomal protein (RILP).Commun Integr Biol. 2014 Oct 31;7(5):e971572. doi: 10.4161/cib.29616. eCollection 2014 Oct. Commun Integr Biol. 2014. PMID: 26843904 Free PMC article. Review.
-
Advances in Use of Capsule-Based Fluorescent Sensors for Measuring Acidification of Endocytic Compartments in Cells with Altered Expression of V-ATPase Subunit V1G1.ACS Appl Mater Interfaces. 2015 Jul 15;7(27):15052-60. doi: 10.1021/acsami.5b04375. Epub 2015 Jul 1. ACS Appl Mater Interfaces. 2015. PMID: 26086317
-
The V-ATPase-inhibitor archazolid abrogates tumor metastasis via inhibition of endocytic activation of the Rho-GTPase Rac1.Cancer Res. 2012 Nov 15;72(22):5976-87. doi: 10.1158/0008-5472.CAN-12-1772. Epub 2012 Sep 17. Cancer Res. 2012. PMID: 22986742
-
Regulation of V-ATPase assembly and function of V-ATPases in tumor cell invasiveness.Biochim Biophys Acta. 2016 Aug;1857(8):1213-1218. doi: 10.1016/j.bbabio.2016.02.010. Epub 2016 Feb 22. Biochim Biophys Acta. 2016. PMID: 26906430 Free PMC article. Review.
Cited by
-
Crosstalk between autophagy inhibitors and endosome-related secretory pathways: a challenge for autophagy-based treatment of solid cancers.Mol Cancer. 2021 Oct 27;20(1):140. doi: 10.1186/s12943-021-01423-6. Mol Cancer. 2021. PMID: 34706732 Free PMC article. Review.
-
Allele-specific silencing as therapy for familial amyotrophic lateral sclerosis caused by the p.G376D TARDBP mutation.Brain Commun. 2022 Dec 16;4(6):fcac315. doi: 10.1093/braincomms/fcac315. eCollection 2022. Brain Commun. 2022. PMID: 36751500 Free PMC article.
-
Autophagy and Lysosomal Functionality in CMT2B Fibroblasts Carrying the RAB7K126R Mutation.Cells. 2022 Jan 31;11(3):496. doi: 10.3390/cells11030496. Cells. 2022. PMID: 35159308 Free PMC article.
-
Induced mitochondrial deficit by NDUFS3 transient silencing reduces RAB7 expression and causes lysosomal dysfunction in pancreatic cancer cells.Cell Commun Signal. 2025 May 14;23(1):224. doi: 10.1186/s12964-025-02214-y. Cell Commun Signal. 2025. PMID: 40369571 Free PMC article.
-
Lysosomal Dysfunction in Amyotrophic Lateral Sclerosis: A Familial Case Linked to the p.G376D TARDBP Mutation.Int J Mol Sci. 2025 Mar 21;26(7):2867. doi: 10.3390/ijms26072867. Int J Mol Sci. 2025. PMID: 40243477 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

