Impact of Chaperone-Mediated Autophagy in Brain Aging: Neurodegenerative Diseases and Glioblastoma
- PMID: 33633561
- PMCID: PMC7901968
- DOI: 10.3389/fnagi.2020.630743
Impact of Chaperone-Mediated Autophagy in Brain Aging: Neurodegenerative Diseases and Glioblastoma
Abstract
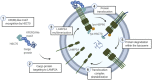
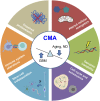
Brain aging is characterized by a time-dependent decline of tissue integrity and function, and it is a major risk for neurodegenerative diseases and brain cancer. Chaperone-mediated autophagy (CMA) is a selective form of autophagy specialized in protein degradation, which is based on the individual translocation of a cargo protein through the lysosomal membrane. Regulation of processes such as proteostasis, cellular energetics, or immune system activity has been associated with CMA, indicating its pivotal role in tissue homeostasis. Since first studies associating Parkinson's disease (PD) to CMA dysfunction, increasing evidence points out that CMA is altered in both physiological and pathological brain aging. In this review article, we summarize the current knowledge regarding the impact of CMA during aging in brain physiopathology, highlighting the role of CMA in neurodegenerative diseases and glioblastoma, the most common and aggressive brain tumor in adults.
Keywords: CMA; LAMP2; glioblastoma; neurodegenerative diseases; physiological aging.
Copyright © 2021 Auzmendi-Iriarte and Matheu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Chaperone-mediated autophagy in neurodegenerative diseases: mechanisms and therapy.Mol Cell Biochem. 2023 Oct;478(10):2173-2190. doi: 10.1007/s11010-022-04640-9. Epub 2023 Jan 25. Mol Cell Biochem. 2023. PMID: 36695937 Review.
-
Chaperone-Mediated Autophagy in Brain Injury: A Double-Edged Sword with Therapeutic Potentials.Mol Neurobiol. 2024 Dec;61(12):10671-10683. doi: 10.1007/s12035-024-04230-4. Epub 2024 May 22. Mol Neurobiol. 2024. PMID: 38775879 Review.
-
Chaperone mediated autophagy to the rescue: A new-fangled target for the treatment of neurodegenerative diseases.Mol Cell Neurosci. 2015 May;66(Pt A):29-36. doi: 10.1016/j.mcn.2015.01.003. Epub 2015 Feb 25. Mol Cell Neurosci. 2015. PMID: 25724482 Review.
-
Chaperone-mediated autophagy: Advances from bench to bedside.Neurobiol Dis. 2019 Feb;122:41-48. doi: 10.1016/j.nbd.2018.05.010. Epub 2018 May 22. Neurobiol Dis. 2019. PMID: 29800676 Review.
-
Chaperone-Mediated Autophagy.Adv Exp Med Biol. 2019;1206:435-452. doi: 10.1007/978-981-15-0602-4_20. Adv Exp Med Biol. 2019. PMID: 31776997 Review.
Cited by
-
Editorial: Protein Misfolding and Proteostasis Impairment in Aging and Neurodegeneration: From Spreading Studies to Therapeutic Approaches.Front Aging Neurosci. 2022 Feb 9;13:830779. doi: 10.3389/fnagi.2021.830779. eCollection 2021. Front Aging Neurosci. 2022. PMID: 35221985 Free PMC article. No abstract available.
-
Lysosomes as a Target of Anticancer Therapy.Int J Mol Sci. 2023 Jan 22;24(3):2176. doi: 10.3390/ijms24032176. Int J Mol Sci. 2023. PMID: 36768500 Free PMC article. Review.
-
Cleavage of Hsp70.1 causes lysosomal cell death under stress conditions.Front Mol Biosci. 2024 May 27;11:1378656. doi: 10.3389/fmolb.2024.1378656. eCollection 2024. Front Mol Biosci. 2024. PMID: 38859931 Free PMC article. Review.
-
The role of SIRT1 in autophagy and drug resistance: unveiling new targets and potential biomarkers in cancer therapy.Front Pharmacol. 2024 Sep 30;15:1469830. doi: 10.3389/fphar.2024.1469830. eCollection 2024. Front Pharmacol. 2024. PMID: 39403142 Free PMC article. Review.
-
Homeostasis of carbohydrates and reactive oxygen species is critically changed in the brain of middle-aged mice: Molecular mechanisms and functional reasons.BBA Adv. 2023 Jan 21;3:100077. doi: 10.1016/j.bbadva.2023.100077. eCollection 2023. BBA Adv. 2023. PMID: 37082254 Free PMC article.
References
-
- Agarraberes F. A., Dice J. F. (2001). A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J. Cell. Sci. 114, 2491–2499. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

