Immune Profiling of Parkinson's Disease Revealed Its Association With a Subset of Infiltrating Cells and Signature Genes
- PMID: 33633562
- PMCID: PMC7899990
- DOI: 10.3389/fnagi.2021.605970
Immune Profiling of Parkinson's Disease Revealed Its Association With a Subset of Infiltrating Cells and Signature Genes
Abstract
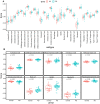
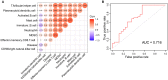
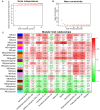
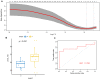
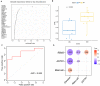
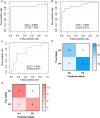
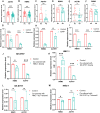
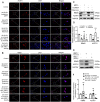
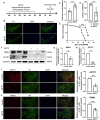
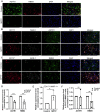
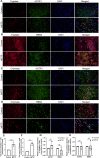
Parkinson's disease (PD) is an age-related and second most common neurodegenerative disorder. In recent years, increasing evidence revealed that peripheral immune cells might be able to infiltrate into brain tissues, which could arouse neuroinflammation and aggravate neurodegeneration. This study aimed to illuminate the landscape of peripheral immune cells and signature genes associated with immune infiltration in PD. Several transcriptomic datasets of substantia nigra (SN) from the Gene Expression Omnibus (GEO) database were separately collected as training cohort, testing cohort, and external validation cohort. The immunoscore of each sample calculated by single-sample gene set enrichment analysis was used to reflect the peripheral immune cell infiltration and to identify the differential immune cell types between PD and healthy participants. According to receiver operating characteristic (ROC) curve analysis, the immunoscore achieved an overall accuracy of the area under the curve (AUC) = 0.883 in the testing cohort, respectively. The immunoscore displayed good performance in the external validation cohort with an AUC of 0.745. The correlation analysis and logistic regression analysis were used to analyze the correlation between immune cells and PD, and mast cell was identified most associated with the occurrence of PD. Additionally, increased mast cells were also observed in our in vivo PD model. Weighted gene co-expression network analysis (WGCNA) was used to selected module genes related to a mast cell. The least absolute shrinkage and selection operator (LASSO) analysis and random-forest analysis were used to analyze module genes, and two hub genes RBM3 and AGTR1 were identified as associated with mast cells in the training cohort. The expression levels of RBM3 and AGTR1 in these cohorts and PD models revealed that these hub genes were significantly downregulated in PD. Moreover, the expression trend of the aforementioned two genes differed in mast cells and dopaminergic (DA) neurons. In conclusion, this study not only exhibited a landscape of immune infiltrating patterns in PD but also identified mast cells and two hub genes associated with the occurrence of PD, which provided potential therapeutic targets for PD patients (PDs).
Keywords: AGTR1; Parkinson’s disease; RBM3; immune cell infiltration; mast cell.
Copyright © 2021 Zhang, Shao, Xu, Liu, Liu, Luo, Jin and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Identifying the hub gene and immune infiltration of Parkinson's disease using bioinformatical methods.Brain Res. 2022 Jun 15;1785:147879. doi: 10.1016/j.brainres.2022.147879. Epub 2022 Mar 10. Brain Res. 2022. PMID: 35278479
-
Parkinson's Disease Gene Biomarkers Screened by the LASSO and SVM Algorithms.Brain Sci. 2023 Jan 20;13(2):175. doi: 10.3390/brainsci13020175. Brain Sci. 2023. PMID: 36831718 Free PMC article.
-
Identification of Immune-Related Hub Genes in Parkinson's Disease.Front Genet. 2022 Jul 22;13:914645. doi: 10.3389/fgene.2022.914645. eCollection 2022. Front Genet. 2022. PMID: 35938039 Free PMC article.
-
Novel diagnostic biomarkers related to immune infiltration in Parkinson's disease by bioinformatics analysis.Front Neurosci. 2023 Jan 26;17:1083928. doi: 10.3389/fnins.2023.1083928. eCollection 2023. Front Neurosci. 2023. PMID: 36777638 Free PMC article.
-
Neuroinflammation in Parkinson's disease and its potential as therapeutic target.Transl Neurodegener. 2015 Oct 12;4:19. doi: 10.1186/s40035-015-0042-0. eCollection 2015. Transl Neurodegener. 2015. PMID: 26464797 Free PMC article. Review.
Cited by
-
Impact of Mast Cell Activation on Neurodegeneration: A Potential Role for Gut-Brain Axis and Helicobacter pylori Infection.Neurol Int. 2024 Dec 6;16(6):1750-1778. doi: 10.3390/neurolint16060127. Neurol Int. 2024. PMID: 39728753 Free PMC article. Review.
-
Single-cell RNA sequencing of CSF reveals neuroprotective RAC1+ NK cells in Parkinson's disease.Front Immunol. 2022 Sep 21;13:992505. doi: 10.3389/fimmu.2022.992505. eCollection 2022. Front Immunol. 2022. PMID: 36211372 Free PMC article.
-
Identification of novel immune-related biomarker and therapeutic drugs in Parkinson disease via integrated bioinformatics analysis.Medicine (Baltimore). 2023 Aug 4;102(31):e34456. doi: 10.1097/MD.0000000000034456. Medicine (Baltimore). 2023. PMID: 37543820 Free PMC article.
-
Identifying crosstalk genetic biomarkers linking a neurodegenerative disease, Parkinson's disease, and periodontitis using integrated bioinformatics analyses.Front Aging Neurosci. 2022 Dec 5;14:1032401. doi: 10.3389/fnagi.2022.1032401. eCollection 2022. Front Aging Neurosci. 2022. PMID: 36545026 Free PMC article.
-
Integrated Bioinformatics Analysis for Revealing CBL is a Potential Diagnosing Biomarker and Related Immune Infiltration in Parkinson's Disease.Int J Gen Med. 2024 May 22;17:2371-2386. doi: 10.2147/IJGM.S456942. eCollection 2024. Int J Gen Med. 2024. PMID: 38799203 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

