Two Novel Endornaviruses Co-infecting a Phytophthora Pathogen of Asparagus officinalis Modulate the Developmental Stages and Fungicide Sensitivities of the Host Oomycete
- PMID: 33633714
- PMCID: PMC7902037
- DOI: 10.3389/fmicb.2021.633502
Two Novel Endornaviruses Co-infecting a Phytophthora Pathogen of Asparagus officinalis Modulate the Developmental Stages and Fungicide Sensitivities of the Host Oomycete
Abstract
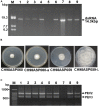
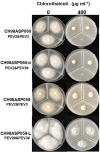
Two novel endornaviruses, Phytophthora endornavirus 2 (PEV2) and Phytophthora endornavirus 3 (PEV3) were found in isolates of a Phytophthora pathogen of asparagus collected in Japan. A molecular phylogenetic analysis indicated that PEV2 and PEV3 belong to the genus Alphaendornavirus. The PEV2 and PEV3 genomes consist of 14,345 and 13,810 bp, and they contain single open reading frames of 4,640 and 4,603 codons, respectively. Their polyproteins contain the conserved domains of an RNA helicase, a UDP-glycosyltransferase, and an RNA-dependent RNA polymerase, which are conserved in other alphaendornaviruses. PEV2 is closely related to Brown algae endornavirus 2, whereas PEV3 is closely related to Phytophthora endornavirus 1 (PEV1), which infects a Phytophthora sp. specific to Douglas fir. PEV2 and PEV3 were detected at high titers in two original Phytophthora sp. isolates, and we found a sub-isolate with low titers of the viruses during subculture. We used the high- and low-titer isolates to evaluate the effects of the viruses on the growth, development, and fungicide sensitivities of the Phytophthora sp. host. The high-titer isolates produced smaller mycelial colonies and much higher numbers of zoosporangia than the low-titer isolate. These results suggest that PEV2 and PEV3 inhibited hyphal growth and stimulated zoosporangium formation. The high-titer isolates were more sensitive than the low-titer isolate to the fungicides benthiavalicarb-isopropyl, famoxadone, and chlorothalonil. In contrast, the high-titer isolates displayed lower sensitivity to the fungicide metalaxyl (an inhibitor of RNA polymerase I) when compared with the low-titer isolate. These results indicate that persistent infection with PEV2 and PEV3 may potentially affect the fungicide sensitivities of the host oomycete.
Keywords: asparagus phytophthora rot fungus; attenuation; endornavirus; fungicide sensitivity; zoosporangium formation; zoospore transmission.
Copyright © 2021 Uchida, Sakuta, Ito, Takahashi, Katayama, Omatsu, Mizutani, Arie, Komatsu, Fukuhara, Uematsu, Okada and Moriyama.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Adams M. J., Lefkowitz E. J., King A. M. Q., Harrach B., Harrison R. L., Knowles N. J., et al. (2017). Changes to taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses. Arch. Virol. 162 2505–2538. 10.1007/s00705-017-3358-5 - DOI - PubMed
-
- Ark P. A., Barret J. T. (1938). Phytophthora rot of asparagus in California. Phytopathology 28 754–756.
LinkOut - more resources
Full Text Sources
Other Literature Sources

